Improved method for identifying target RNA sequences of RNA-binding protein in cell sample
A technology that binds proteins and targets, applied in the biological field, can solve problems such as isotope hazards, cumbersome steps, and complicated processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
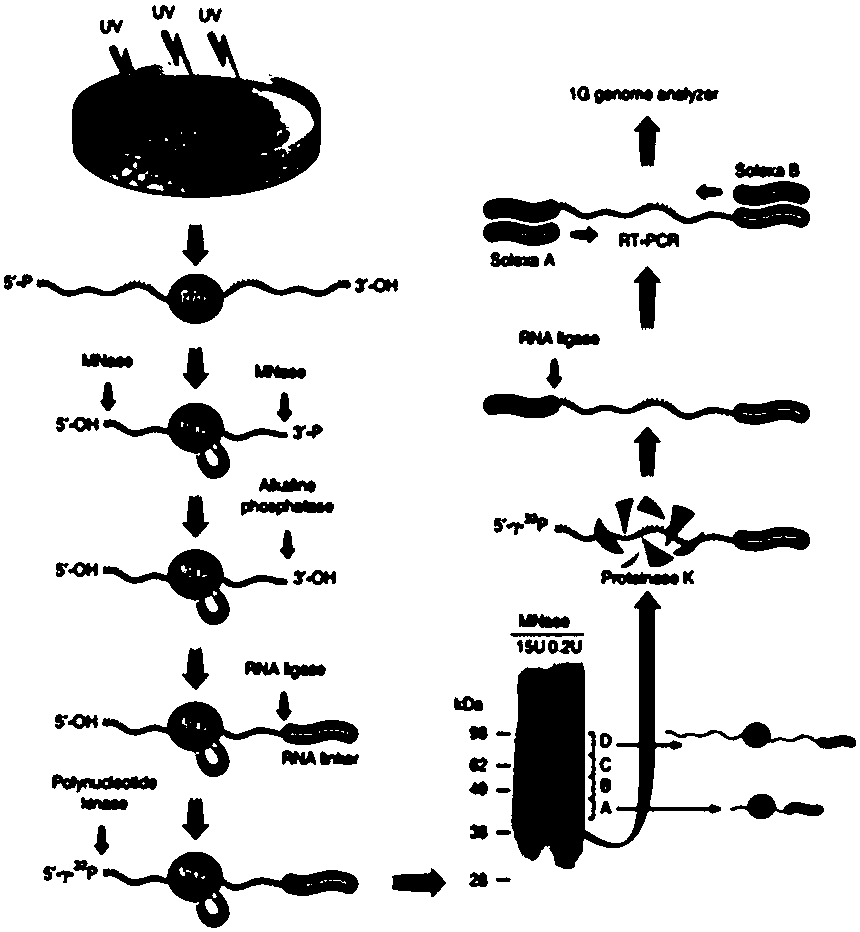
[0053] [Example 1] Design and synthesis of RNA linker
[0054] The linker sequence used matches illumina's Nex-500 sequencer:
[0055] 3'linker: 5'-PO 4 ATC TGG AAT TCT CGG GTG CCA AGG-Puromycin 3' (Seq ID NO: 1);
[0056] 5'linker: 5'Biotin-TGG AAT TCT CGG GTG CCA AGG 3'(Seq ID NO:2);
[0057] The puromycin at the 3' end of the 3'linker and the biotin at the 5' end of the 5'linker block the linker end to prevent head-to-tail self-connection.
[0058] The reverse transcription primer is the reverse complementary DNA sequence of the 3' linker:
[0059] 5'CCT TGG CAC CCG AGA ATT C (Seq ID NO:3),
[0060] Forward primers used in PCR:
[0061] 5'AAT GAT ACG GCG ACC ACC GAC AGG TTC AGA GTT CTA CAG TCC GA (Seq ID NO:4);
[0062] The reverse primer is the reverse transcription primer.
Embodiment 2
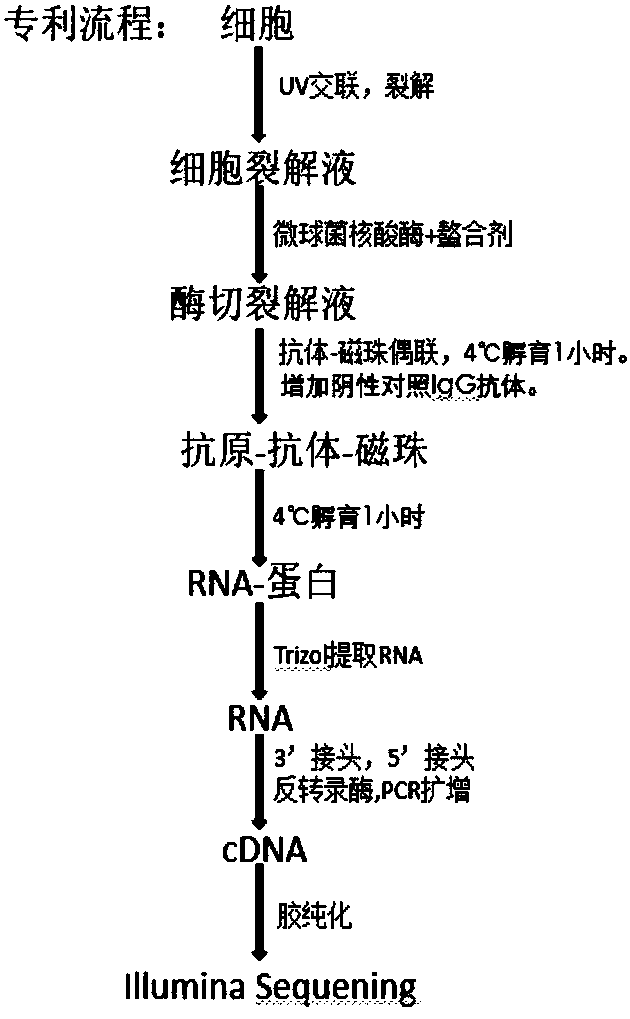
[0063] [Example 2] Preparation of ultraviolet crosslinking and cell lysate
[0064] 1) Use a 150mm cell culture dish to culture HeLa cells, use DMEM (Invitrogen) as the medium, discard the medium when the cell abundance reaches over 80%, rinse twice with 1xPBS buffer, then add an appropriate amount of PBS buffer to cover cell surface;
[0065] 2) Put the petri dish on ice, put it into the drawer at the lower part of HL-2000 crosslinking instrument (MVP), make 400mj / cm 2 Energy of 254nm ultraviolet radiation 1 time. Then take out the culture dish, pour off the PBS, add 15ml of PBS, and scrape off the cells with a cell scraper;
[0066] 3) Use a pipette to suck the PBS suspension of all cells into a 15ml centrifuge tube, centrifuge at 4000rpm at 4°C for 5 minutes, and remove the supernatant. Add 1.5ml PBS, blow up the precipitate with a pipette, transfer to a clean 1.5ml centrifuge tube (DEPC treatment or use imported RNase free centrifuge tube), centrifuge again at 4000rpm f...
Embodiment 3
[0072] [Example 3] Fragmentation of RNA molecules
[0073]When performing a RIP experiment on a protein for the first time, it is necessary to explore the optimal concentration of MicrococalNμclease enzyme (MNase) (Fermentas company) so that the RNA tag is at least 20 nucleotides (20 nucleotides are MCSC Genome Browser for genome comparison (minimum length), the average length is preferably 30-60 nucleotides.
[0074] 1) Add Micrococal Nμclease enzyme and 10X MNase buffer according to the explored concentration, put it on the thermomixer for 10 minutes, 1000rpm, and mix for 15 seconds every 3 minutes;
[0075] 2) Add EDTA+EGTA solution to chelate Ca2+ in the system to terminate the reaction.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com