SSR molecular marker primer capable of rapidly identifying robinia pseudoacacia polyploids
A technology of molecular markers and polyploids, applied in the determination/testing of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem of rapid identification of Robinia pseudoacacia polyploids, which cannot be quickly identified High-efficiency SSR primers and other issues to achieve high polymorphism and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The SSR molecular marker primers for rapid identification of Robinia pseudoacacia polyploids were obtained through the following steps:
[0029] (1) Extraction of Robinia pseudoacacia total RNA and transcriptome sequencing
[0030] Total RNA was extracted using Trizol reagent. The pipette tip, mortar and spoon used in the experiment were all treated with DEPC (diethyl pyrocarbonate) for 4 hours, followed by high temperature and high pressure sterilization for 1 hour. Then, total RNA was extracted according to the usage method of Trizol reagent. Use 1.0% non-denaturing agarose gel electrophoresis to roughly detect the integrity of the extracted RNA; use the instrument Nanodrop (IMPLEN, CA, USA) to detect the purity and concentration of the extracted RNA (OD 260 / 230 ratio; OD 260 / 280 ratio) ; RNA integrity was detected using Agilent 2100 (Agilent Technologies, CA, USA).
[0031] After the RNA sample is qualified, in order to ensure high-quality transcriptome data, it w...
Embodiment 2
[0048] 1. Test sample
[0049] In this experiment, five tetraploid Robinia pseudoacacia clones introduced from Korea were used as test materials. They are K1 (Tetraploid R. pseudoacacia K1), K2 (Tetraploid R. pseudoacacia K2), K3 (Tetraploid R. pseudoacacia K3), K4 (Tetraploid R. pseudoacacia K4), and K5 (Tetraploid R. pseudoacacia K5).
[0050] The materials were collected from Daqingshan Forest Farm, Fei County, Shandong Province.
[0051] In May 2016, the young leaves were collected, dried on silica gel, and set aside.
[0052] 2. Genomic DNA extraction:
[0053] Take the young leaves of Robinia pseudoacacia clones, and use the plant tissue genomic DNA extraction kit (magnetic bead method) (Cat. No. PTED-6030) of Inrich Biochemical Technology (Shanghai) Co., Ltd. to extract according to the instructions.
[0054] 3. Use SSR primers for PCR amplification:
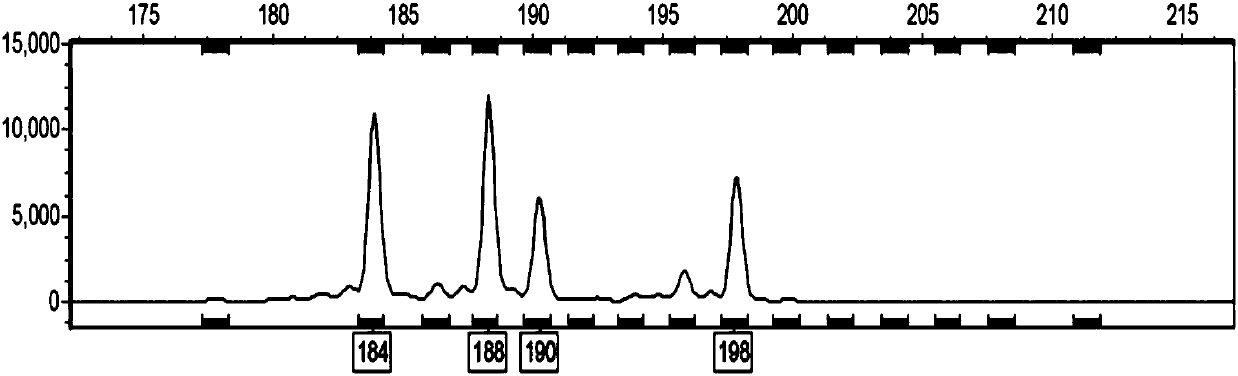
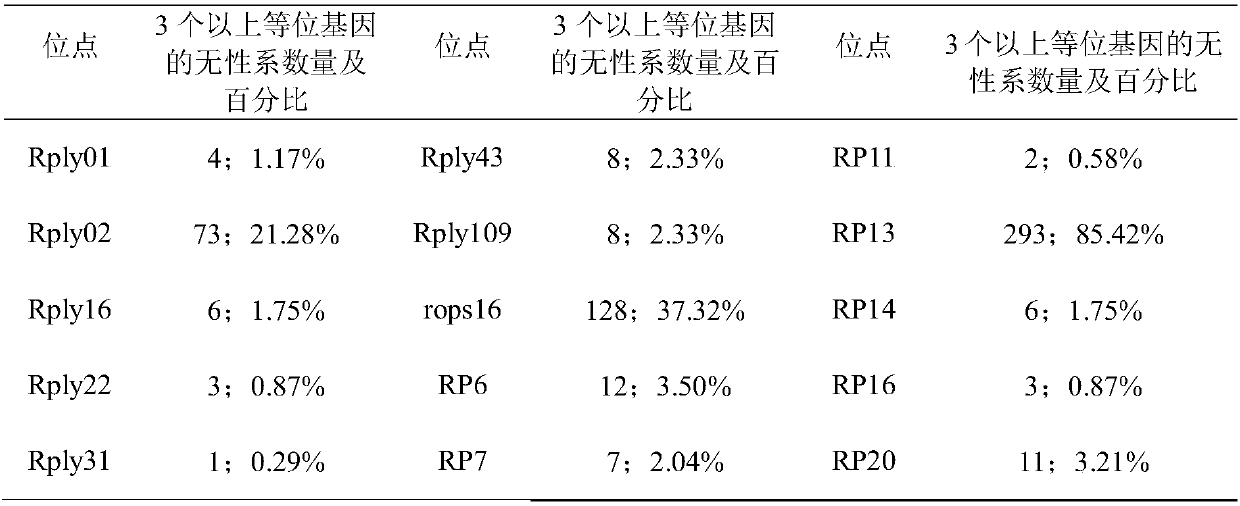
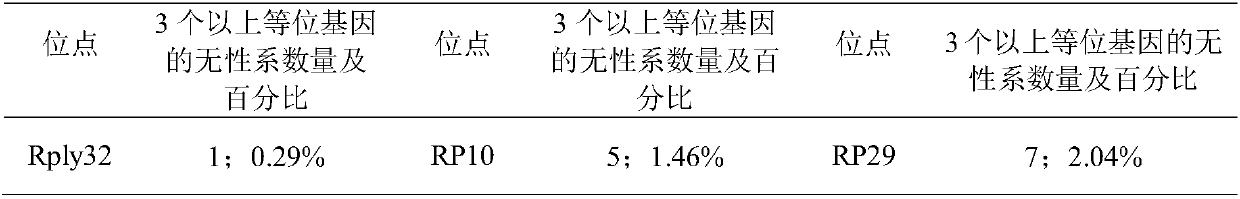
[0055] Using the total DNA sample of Robinia pseudoacacia extracted in step 2 as a template, use the following two ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com