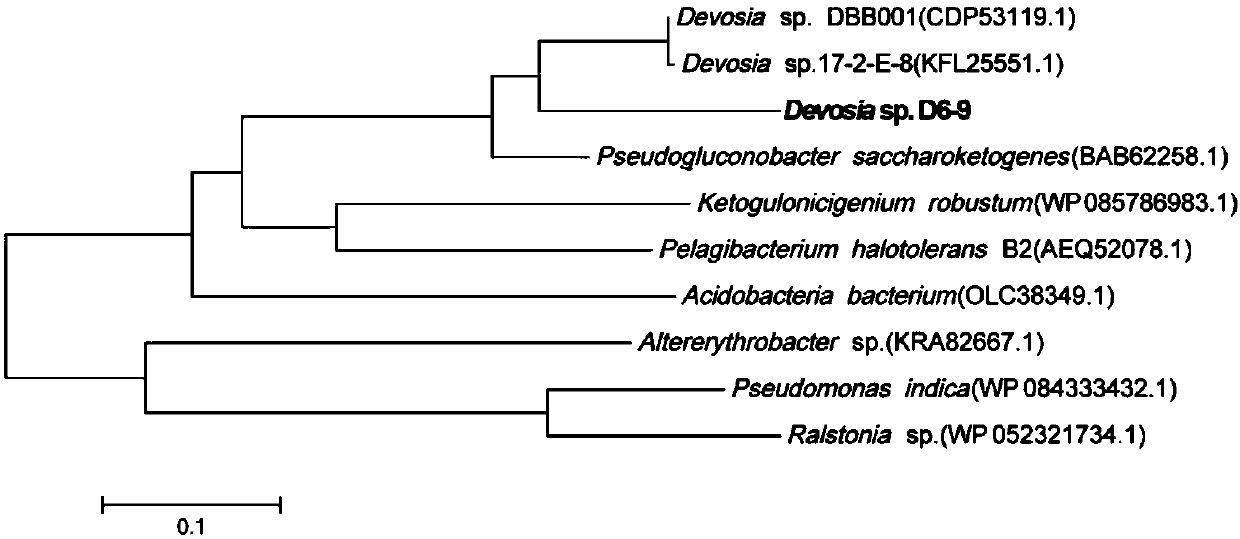
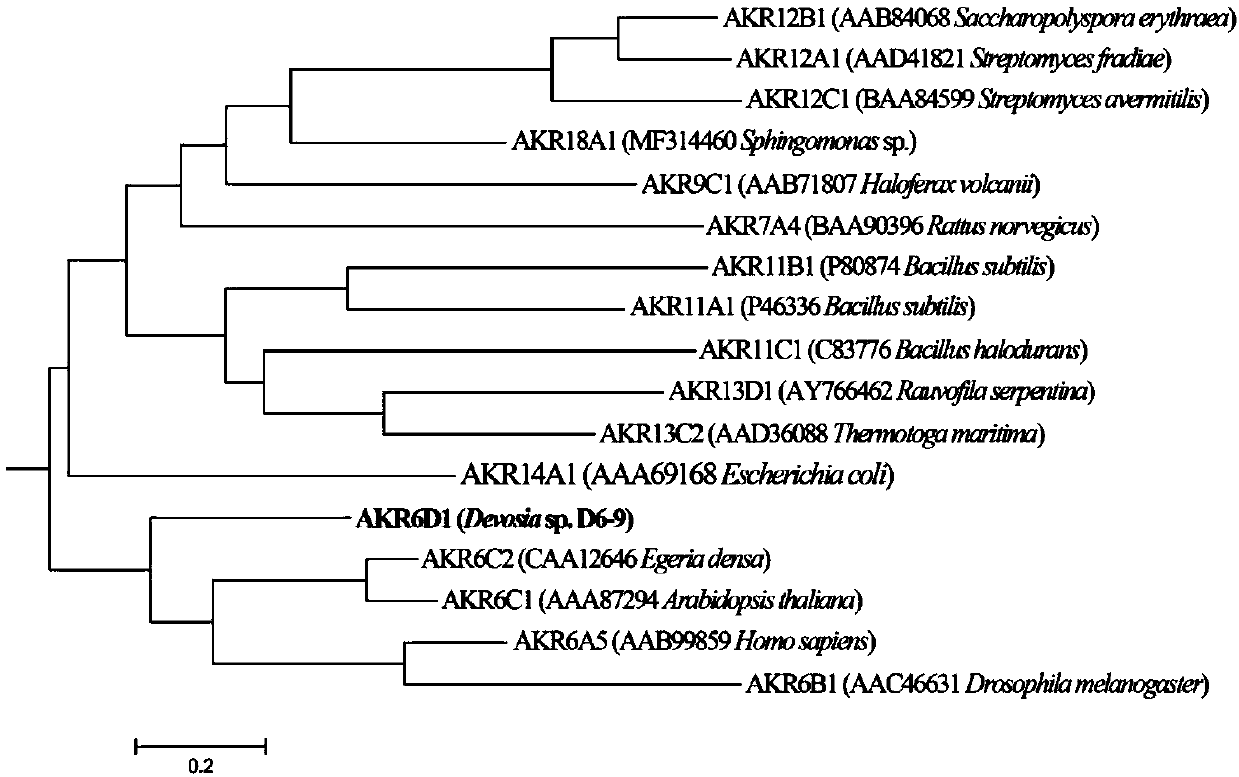
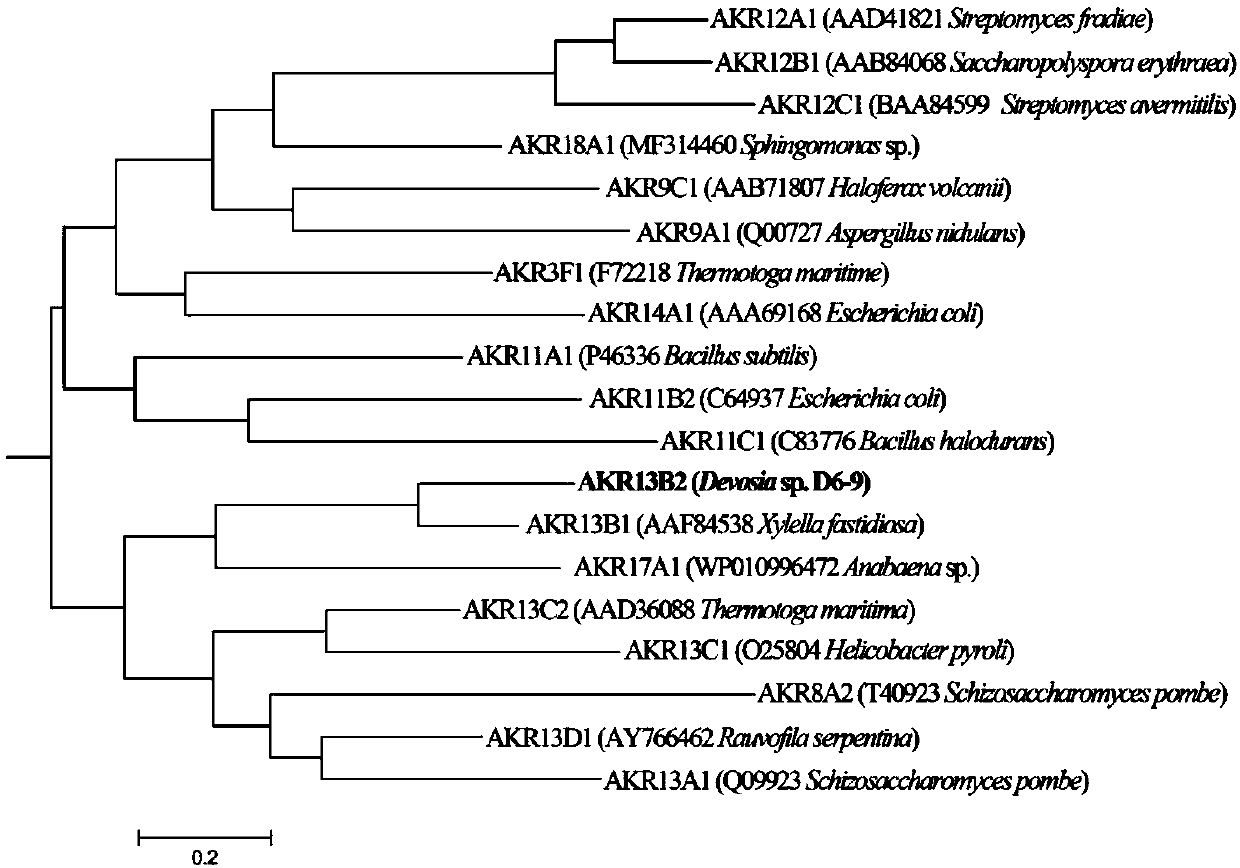
Fusarium toxin removal path related genes ADH, AKR6D1 and AKR13B2 and their application
A fusarium and gene technology, applied in application, genetic engineering, plant genetic improvement, etc., can solve the problem of unknown genes related to metabolic pathways, and achieve the effects of product specificity, huge development and application potential, and simple cloning operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Cloning of target genes ADH, AKR6D1, AKR13B2 and construction and transformation of prokaryotic expression vectors
[0029] Target sequence cloning: Using the genomic DNA of Devosia sp. degrading bacteria D6-9 (provided by the research group laboratory) as a template, the high-fidelity enzyme KOD plus (purchased from Toyobo, Japan) was used to amplify the target sequence Gene fragment. Amplification primer of gene ADH, forward primer: 5′-CATGCTGACGGGGCCGCGG-3′, reverse primer is 5′-CTTGGCTTCGGGCAGGGCG-3′; amplification primer of gene AKR6D1, forward primer is 5′-ATGGAATATCGTCGTCTGGG-3′, The reverse primer is 5′-CTAGAACCGCTGCGGGCCGG-3′; the amplification primer of the gene AKR13B2, the forward primer is 5′-ATGTCAGAACCCAATGCAGC-3′, and the reverse primer is 5′-TCACGAAGCCCCTTTCCAGCTC-3′. 50μL reaction system: 5μL of 10×KOD buffer, 25mmol / L MgSO 4 2 μL, 5 μL of 2 mmol / L dNTPs, 1.5 μL of each 10 μmol / L primer, 1 μL of KOD plus (1U / μL), 1 μL of template cDNA, 5 μ...
Embodiment 2
[0032] Example 2: Expression and purification of ADH, AKR6D1, AKR13B2 proteins
[0033] Induced expression of protein: culture the Escherichia coli BL21 strain containing the expression vector in LB liquid medium, in a shaker at 37°C (200r / min) to OD 600 It reaches about 0.6; add IPTG (purchased from Sigma, USA) at a final concentration of 0.4 mM, place in a shaker at 16°C (140r / min) for induction and culture for 12h.
[0034] Purification of protein: collect the cells by centrifugation, and resuspend the cells with an appropriate amount of lysis buffer (according to the ratio of 1:20-1:40). Vortex vigorously to fully dissolve the cells. Before cell disruption, 100 μM protease inhibitor PMSF was added at a ratio of 1:100 (v:v), and E. coli cells were disrupted with a high-pressure cell disruptor. The bacterial cell disruption solution was centrifuged at 16000r / min for 30min, the supernatant was collected, and all passed through the protein purification column filled with Ni-...
Embodiment 3
[0035] Embodiment 3: the catalysis of purified ADH protein to DON
[0036] The purified ADH protein was reacted with DON in buffer, in a 50 μL reaction system, including 6 μg purified protein, 100 μM DON, 100 μM PQQ, 100 μM Ca + , add Tris-HCL buffer solution (pH 7.5) to a total volume of 50 μL, and react at 30°C. After the reaction is completed, add an equal volume of methanol to end the reaction. Use high-performance liquid chromatography (HPLC) to detect the DON content in the solution, and analyze the composition of the system by HPLC : Agilent 1200 semi-preparative HPLC main components: quaternary pump (1260QuatPump VL), autosampler (1260ALS), column thermostat (1260FCC), UV detector (1260VWD), automatic fraction collector (1260FC-AS ), analytical chromatographic column (Eclipse XDB-C18, 4.6×150mm, 5μm), semi-preparative chromatographic column (EclipseXDB-C18, 9.4×250mm, 5μm), operating system (Agilent ChemStation, B.04.03). HPLC analysis conditions: injection volume 10μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com