Special CAPS marker for identifying wild type or mutant paddy rice salt resistant gene OSRR22, primer, and primer applications
A salt-tolerant gene and wild-type technology, applied in the field of plant biology, can solve problems such as lack of salt tolerance, achieve the effects of reducing costs, improving breeding efficiency, and simple sequencing methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] This example relates to the creation of a rice salt tolerance gene Osrr22 deletion mutant based on CRISPR-Cas9 technology.
[0036] (1) Guide RNA target sequence selection and primer design
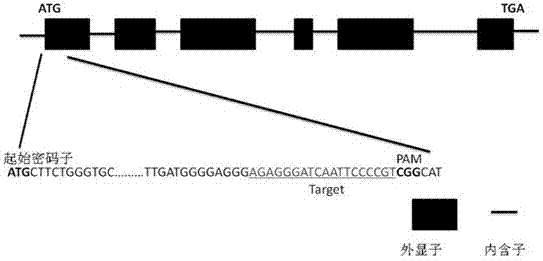
[0037] According to the genome sequence of the control rice salt tolerance gene OsRR22 (GenBank: BR000251.1), an sgRNA of the salt tolerance gene OsRR22 was designed. The 20nt nucleotide sgRNA target sequence is designed according to the 5'-N20-NGG-3' sequence, and the target sequence primers RR22-gRT+ and RR22-OsU6aT- are designed at the same time. Paired with the U6a promoter. Compare the designed sgRNA target sequence to the rice genome database to exclude non-specific target cutting sites. The specific target nucleotide sequence is as follows, see figure 1 .
[0038] RR22-gRT+: 5’-AGAGGGATCAATTCCCCGTgttttagagctagaaat-3’
[0039] RR22-OsU6aT-:5'-ACGGGGAATTGATCCCTCTCggcagccaagccagca-3'
[0040] (2) Construction of target sequence sgRNA expression cassette
[0041] Referring t...
Embodiment 2
[0070] This example relates to the establishment of specific CAPS markers for the identification of rice salt tolerance gene OsRR22 wild type and mutant
[0071] (1) Primer design
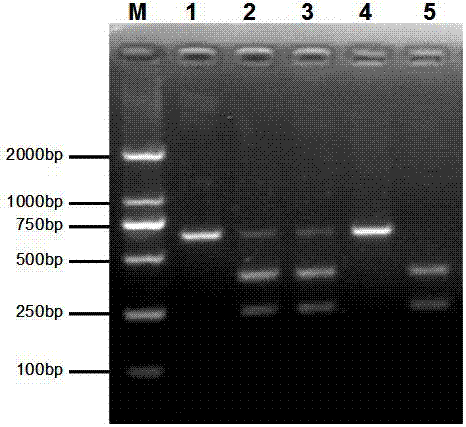
[0072] The rice salt tolerance gene OsRR22 was amplified and sequenced in the japonica rice line WDR58 and the WDR58-cas-1 mutant, and the analysis found that the WDR58-cas-1 mutant had 1 bp at the 69th nucleotide position of the Osrr22 gene locus (A) The insertion leads to amino acid frameshift mutation, premature translation termination, resulting in loss of OsRR22 gene function and thus improving salt tolerance. Bioinformatic analysis of this region revealed that the 1bp insertion generated a Tail I restriction endonuclease site (5'...ACGT⊥...3'), while the wild type did not contain a Tail I restriction endonuclease site point(5'...CGT...3'), such as figure 2 . The upstream and downstream primers CAPS-TailⅠ containing this site were designed on both sides of the 1bp insertion mutation region...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com