Application of Natural Small Molecular Compounds in Inhibiting Ubiquitin Chain Synthesis
A technology for small molecule compounds and synthesis reactions, applied in the fields of biotechnology and medicine, it can solve the problems of unsatisfactory specificity, high cytotoxicity, and many cell signaling pathways, and achieve the effect of good specificity, low IC50 value and high efficacy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Preparation of each raw material in embodiment 1 ubiquitin chain synthesis reaction
[0074] 1. Purification of ubiquitin (Ub)
[0075] Untagged ubiquitin cDNA was constructed on the prokaryotic expression vector pET14b. transforming Escherichia coli
[0076] (BL21(DE3) / pJY2 strain), expression was induced with 0.5 mM IPTG for 4 hours at 37°C. After collecting 1L of Escherichia coli and discarding the supernatant, resuspend with 20ml of buffer solution (50mM Tris-Cl pH7.6, 1mM PMSF, 0.02% (v / v) NP-40, 0.4mg / ml lysozyme), and carry out Sonication. After centrifugation at 20,000g for 20 minutes, transfer the supernatant to a clean container, and slowly add 70% perchloric acid dropwise while stirring at 4°C. At this time, most of the miscellaneous proteins will denature and precipitate, and continue stirring when the pH reaches about 4.0. After 30 minutes, centrifuge at 20000 g for 20 minutes, and take the supernatant. Dialyze twice against 100 volumes of pH 4.5 ammon...
Embodiment 2
[0085] 1. Ubiquitin chain synthesis reaction
[0086] Prepare the following reaction system:
[0087]
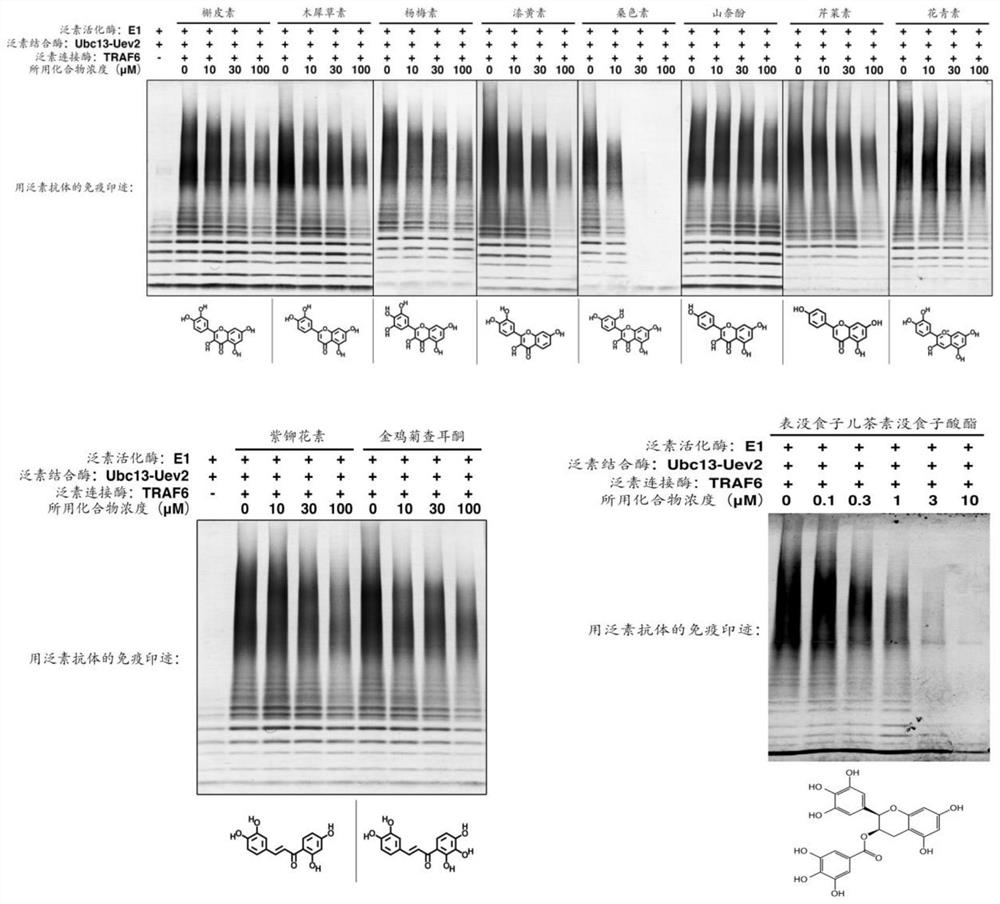
[0088]A control group and 11 sample groups were set; wherein, the control group also contained 5% DMSO, and the sample group also contained natural small molecule compounds (purchased from Selleck, dissolved in DMSO), and the natural small molecule compounds were quercetin ( Sample group 1), luteolin (sample group 2), myricetin (sample group 3), fisetin (sample group 4), morin (sample group 5), kaempferol (sample group 6), apigenin ( Sample group 7), anthocyanin (sample group 8), buterin (sample group 9), coreopsis chalcone (sample group 10) and epigallocatechin gallate (sample group 11).
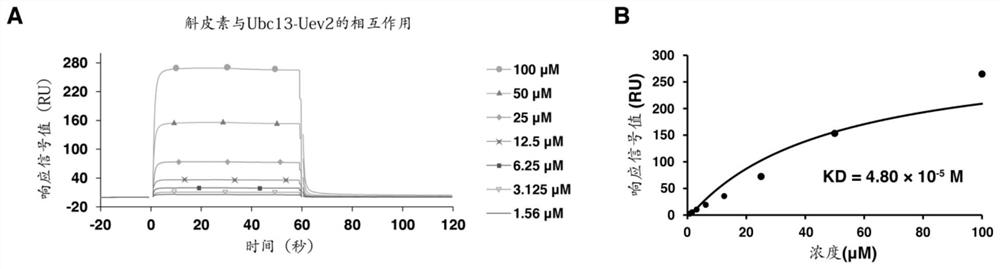
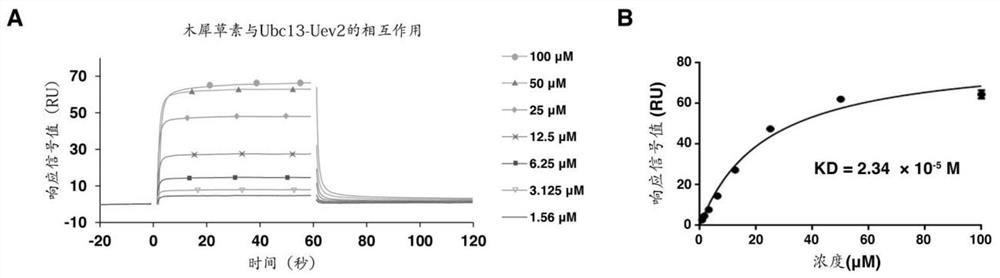
[0089] After the sample group and the control group were reacted at 30°C for 30 minutes, SDS-PAGE sample buffer was added to terminate the reaction, and then electrophoresis and Western Blot were performed to detect ubiquitin protein. The detection results were as follows: figure 1 s...
Embodiment 3
[0097] 1. The experiment of synthesizing ubiquitin chains by using in vitro ubiquitin aminolysis reaction shows that natural small molecule compounds inhibit the enzymatic activity of Ubc13
[0098] Prepare the following reaction system:
[0099]
[0100] Control group and sample group are set; Wherein, control group also contains 5% DMSO, and sample group also contains natural small molecule compound (dissolved in DMSO), and this natural small molecule compound is quercetin (sample group 1), luteolin (sample group 2), epigallocatechin gallate (sample group 3). The same group of samples was set under two conditions of adding or not adding 10nM TRAF6 protein, and placed at 30°C for a reaction ranging from 5 minutes to 4 hours. After the reaction products were separated by SDS-PAGE electrophoresis, the ubiquitin protein was detected by WesternBlot. Results such as Figure 9 ~ Figure 11 shown.
[0101] Depend on Figures 9 to 11 As shown, it shows that dendrobetin, luteoli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com