Specific primer, probe and real-time fluorescence quantitative PCR (Polymerase Chain Reaction) kit for detecting cronobacter sakazakii
A technology of real-time fluorescence quantification and Enterobacter sakazakii, applied in the field of molecular biology and in vitro diagnostic reagents, can solve problems such as unfavorable, finding pathogens, and a large amount of time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 116
[0029] The preparation of embodiment 1 16SrDNA gene standard
[0030] To establish a real-time fluorescent quantitative PCR method, the external standard required by the method must first be prepared. The standard should contain highly conserved and specific sequences, and high specificity of the reaction must be ensured. The 16S rDNA gene widely exists in Enterobacter sakazakii and has high conservation. The present invention uses Enterobacter sakazakii 16SrDNA gene as the target sequence. This example mainly uses PCR technology to amplify the 16SrDNA gene of Enterobacter sakazakii, and uses gene recombination technology to connect it to the plasmid vector pMD18-T to construct the recombinant plasmid pMD18-T-16S, and carry out corresponding PCR identification and sequencing identification , and finally quantified as a standard for the method to be established, laying the foundation for the next method and evaluation.
[0031] 1. Preparation of template DNA
[0032] Genomic...
Embodiment 2
[0095] Embodiment 2 real-time fluorescent quantitative PCR kit
[0096] 1. Design and synthesis of specific primers and probes
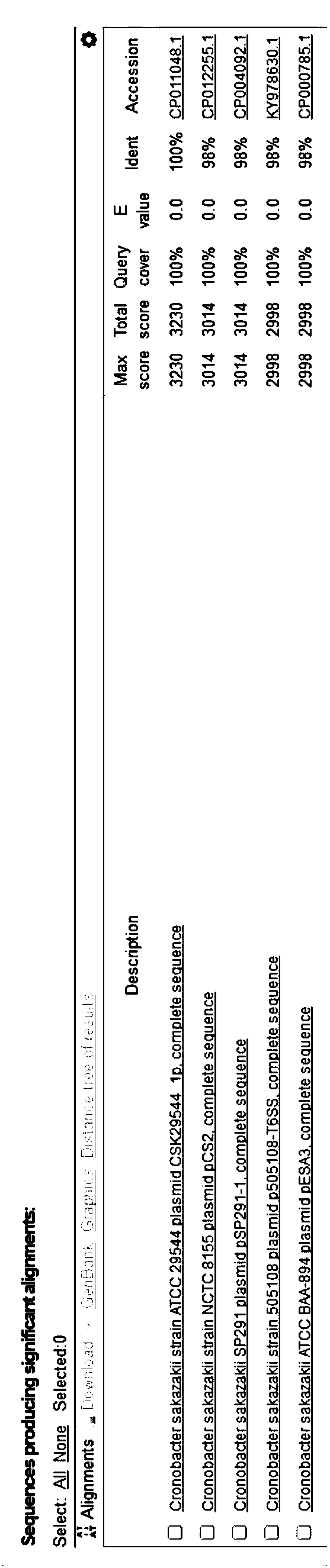
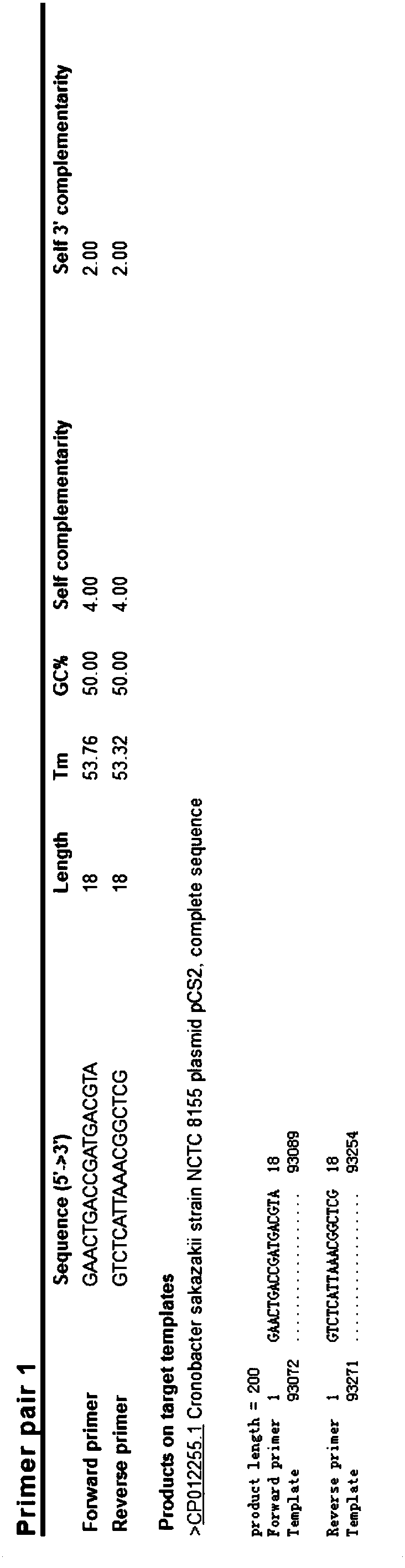
[0097] The present invention performs bioinformatics comparison and analysis on the complete sequence of Enterobacter sakazakii 16SrDNA gene in the NCBI database, selects the conserved fragment sequence suitable for designing primers and probes as the target (see Example 1), and further applies Primer express 3 Software, Primer Premier 5 software and Oligo 7 software, designed multiple sets of real-time fluorescent quantitative PCR primers and probes, after preliminary screening of the test, finally determined a set of fluorescent quantitative PCR primers and probes for the detection of Enterobacter sakazakii.
[0098] As the core of the present invention, a group of primers and probe nucleotide sequences for real-time fluorescent PCR detection of Enterobacter sakazakii are as follows:
[0099] Upstream primer: 16SrDNA-304F 5'-GAACTGACCGATGACGTA-3' ...
Embodiment 3
[0119] Example 3 The quantitative detection method of the sample to be tested by the real-time fluorescent quantitative PCR kit
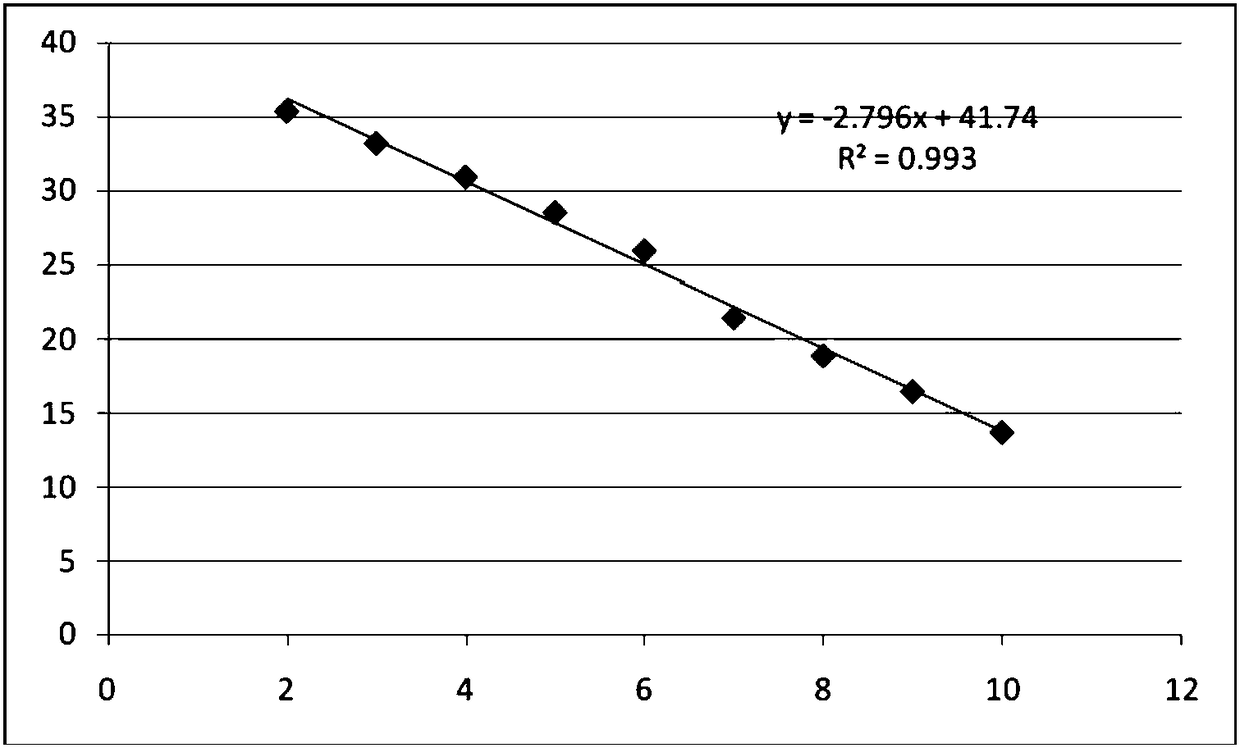
[0120] Respectively with the 16SrDNA gene series concentration standard substance (series concentration standard substance serial concentration of sample DNA to be tested and embodiment 1 preparation is: 1.00 * 10 10 copies / ml, 1.00×10 9 copies / ml, 1.00×10 8 copies / ml, 1.00×10 7 copies / ml, 1.00×10 6 copies / ml, 1.00×10 5 copies / ml, 1.00×10 4 copies / ml, 1.00×10 3 copies / ml, 1.00×10 2 copies / ml) as a template, use the primers and probes in the kit to perform fluorescent quantitative PCR amplification, and set up positive and negative controls at the same time.
[0121] The PCR reaction system is as follows:
[0122]
[0123] Reaction conditions: 94°C pre-denaturation for 2 minutes, 94°C for 10 seconds, 60°C for 30s and collecting fluorescence signals, 40 cycles. Draw a standard curve, and perform rapid quantitative detection through the sta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com