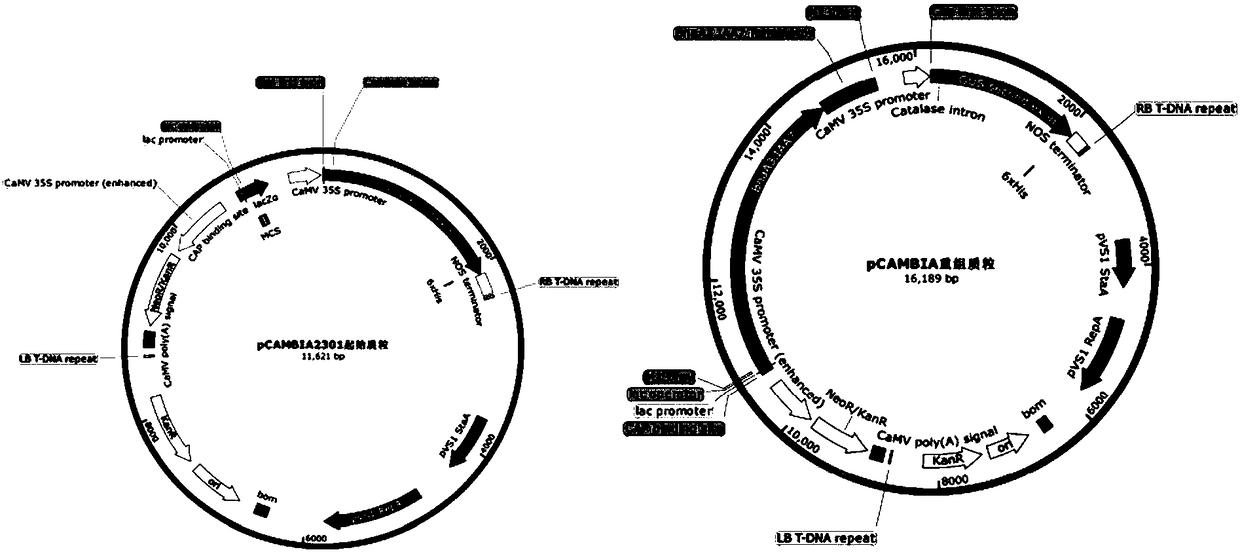
Gene for controlling rape plant type, molecular marker and application thereof
A technology of molecular markers and genes, applied in application, genetic engineering, plant genetic improvement, etc., can solve problems such as unclear application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
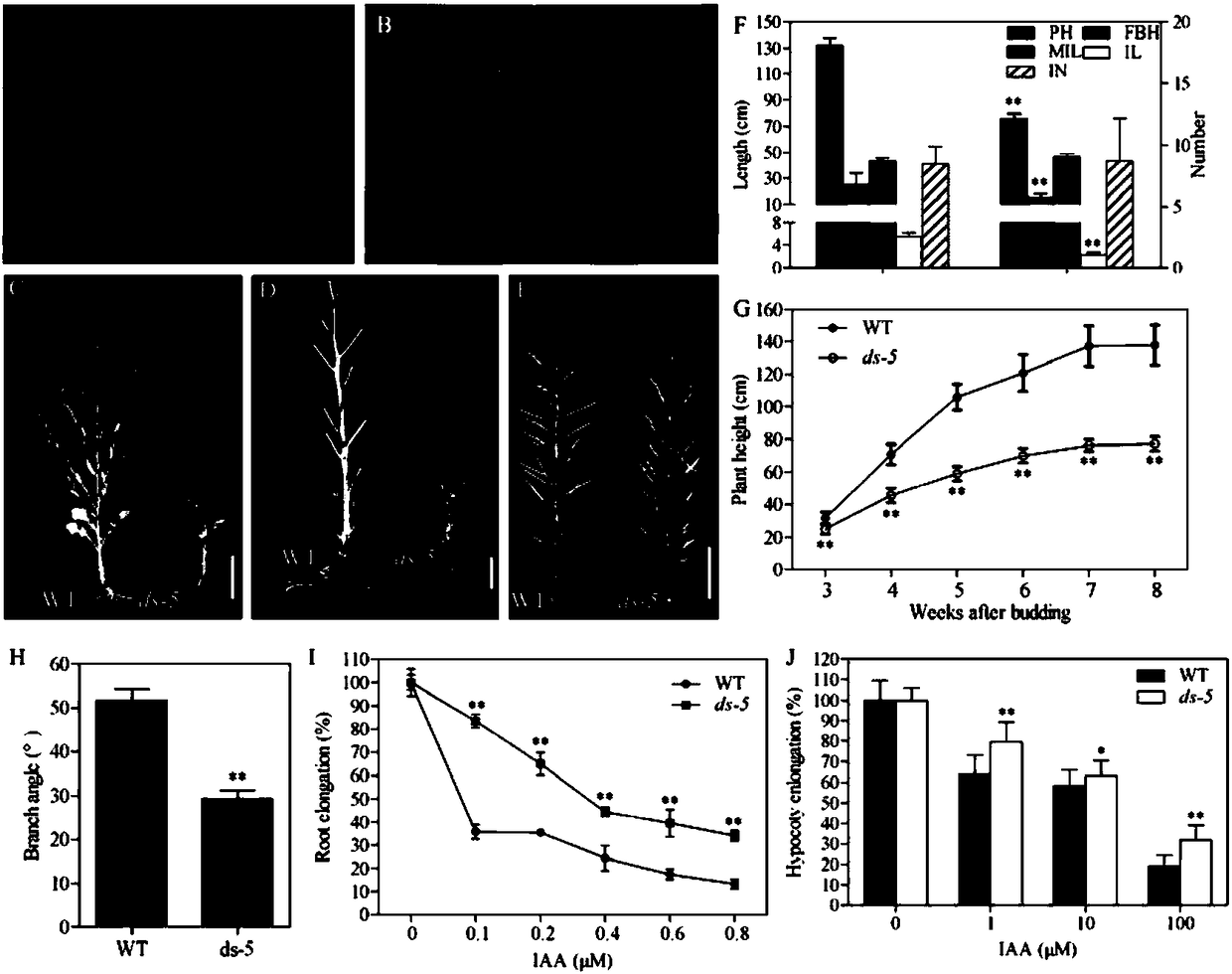
[0063] Example 1: Phenotype identification of Brassica napus mutant ds-5
[0064] (1) The wild type of Brassica napus (remarks in this manual: the "wild type" mentioned in the present invention refers to non-transgenic material refers to the original variety) ds-5 seeds and the Brassica napus mutant ds-5 The seeds (preservation number is CCTCC NO: P201721) were soaked in 70% alcohol for 1 minute, then sterilized in 50% sodium hypochlorite solution for 15 minutes, and finally washed 3 times with sterile water. Sow the treated seeds in a box filled with MS medium (MS+1% agar gel), and observe the length of the hypocotyl after cultivating for one week at 24°C (16h light / 8h dark), the results are as follows: figure 2 As shown in panel A in the figure: the hypocotyl of the mutant is dark green and obviously shortened.
[0065] (2) The wild-type ds-5 and mutant ds-5 were planted in the experimental field of Huazhong Agricultural University, and the phenotypes of the two were obser...
Embodiment 2
[0067] Embodiment 2: Map-position cloning of DS5 gene
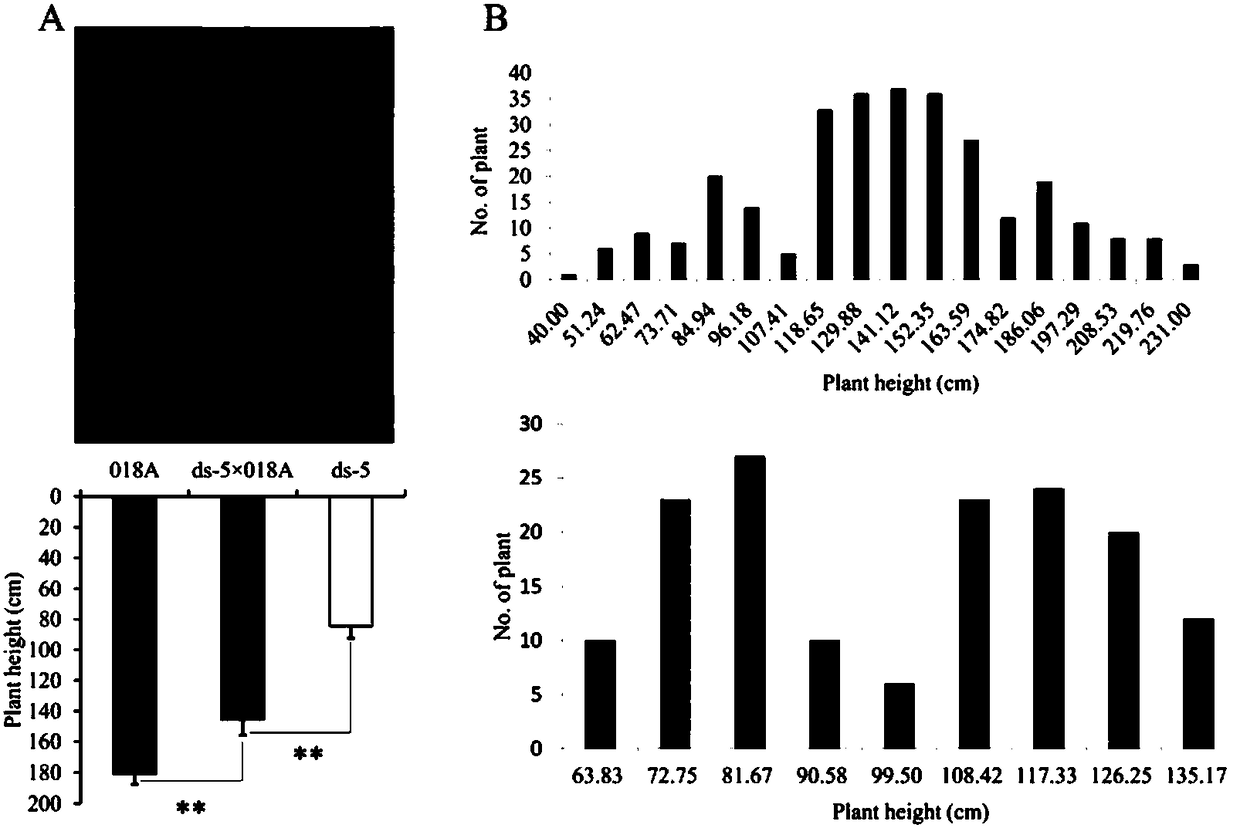
[0068] (1) F 1 generation. f 1 F 2 generation segregation population, and simultaneously crossed with ds-5 to obtain BC 1 Separate groups. Subsequently, for F 2 Generations and BC 1 Each individual plant of the generation was bagged and selfed to obtain F 2:3 and BC 1:2 family lineage. The two parents and F1, F 2 and BC 1 segregating populations and F 2:3 and BC 1:2 The families were planted in the experimental field of Huazhong Agricultural University, and the plant height was investigated and measured at the mature stage. The result is as image 3 Shown, F 1 The plant height of the generation was between the parents and had a significant difference (P2 The plant height of a single plant in the population is between 40cm-231cm and presents a trimodal distribution; F 2:3 The plant height test of the offspring showed that F 2 The population consisted of 69 homozygous short stems, 157 heterozygous intermedi...
Embodiment 3
[0076] Example 3: Development and detection of allele-specific markers
[0077] (1) For the SNP located in the second exon (G>A), use dCAPS Finder 2.0 (http: / / helix.wustl.edu / dcaps / ) to design an allele-specific primer AS_dCPAS, whose sequence is AS_dCPAS-F:TTAAGCATATCCATAACTTTGAAATTAAAT, AS_dCPAS-R:GAGTCATCATGTTCTTCCTGTAGTTCCTCACAGGTGGGAAT( Figure 6 in A panel).
[0078] (2) The detection method of AS_dCPAS marker is as follows: firstly carry out PCR reaction, 20μl PCR system includes: 75ngDNA template, 2μl 10×Taq buffer, 1.6μl MgCl 2 (25mM), 0.4μl dNTPs (10mM), 2μl PCRprimers (1μl each of forward and reverse primers, the concentration is 10μM each) and 5U Taq DNApolymerase (recombinat, Fermentas), supplemented with ddH 2 0 to 20 μl. Reaction program: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 30 sec, renaturation at 65°C for 30 sec, extension at 72°C for 40 sec, a total of 40 cycles; extension at 72°C for 5 min, storage at 4°C. Then carry out enzyme di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com