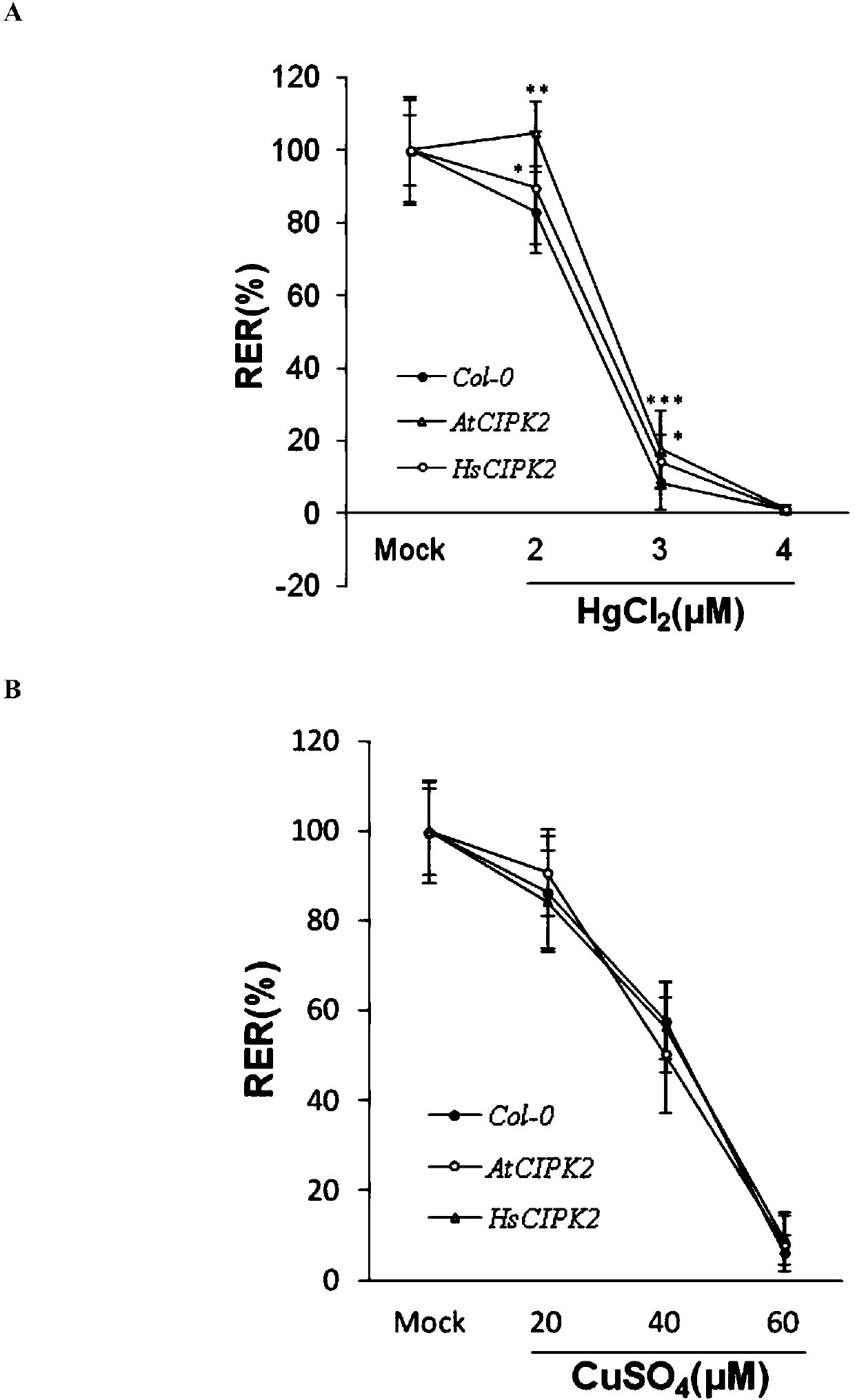
Application of CIPK2 to improvement of mercury resistance/tolerance ability of rice
A kind of use and tolerance technology, applied in the field of plant genetic engineering, can solve the problems of heavy metal stress and other problems, achieve good application prospects, promote growth and development, resist adversity stress or increase yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Embodiment 1, the cultivation of Arabidopsis thaliana, Qinghai-Tibet Plateau annual wild barley and rice
[0068] Arabidopsis wild-type (Col-0) seeds were surface-sterilized (disinfected with 70% ethanol for 10 minutes, then sterilized with 10% NaClO for 30 minutes, and finally rinsed with water for 8 times), cold-treated at 4°C for 3 days in the dark, and placed in 0.5 ×Murashige&Skoog (MS) agar plate (1.5% agar and 1% sucrose, W / V, pH 5.6) cultured. Seedlings were grown in artificial climate culture room (day and night temperature 22 / 20°C, 16 / 8 hour photoperiod, light intensity 80 μmol m -2 the s -1 ), the agar plate was placed vertically.
[0069] The annual wild barley X74 (Hordeum spontaneum C.Koch) and rice Nipponbare (Oryza sativa L.ssp.japonica) seeds on the Qinghai-Tibet Plateau were surface-sterilized with 70% ethanol for 10 minutes, followed by 10% NaClO for 30 minutes, and finally rinsed with water for 8 times . In the dark at 25°C, sterile barley seeds ...
Embodiment 2
[0070] Cloning of embodiment 2, Arabidopsis AtCIPK2 and wild barley HsCIPK2 gene
[0071] The full-length cDNAs of ATCIPK2 (AAF86506; AT5G07070) and OsCIPK2 (ACD7694) were used as probes in the full-length cDNA library of barley (Hordeum vulgare L.; http: / / earth.lab.nig.ac.jp / ~dclust / cgi- bin / barley_pub / ) to search for homologous genes. Based on the highly homologous cDNA sequences to ATCIPK2 and OsCIPK2, the full-length coding sequence (CDS) of HSCIPK2 was amplified by reverse transcription (RT)-PCR. The total RNA of annual wild barley X74 on the Qinghai-Tibet Plateau was isolated with RNeasy Plant Mini Kit (QiAGEN), and then the first-strand cDNA was synthesized with SuperScript III First-Strand Synthesis System (Invitrogen). The amino acid alignment of CIPK2 was analyzed by bioinformatics using Lasergene software (MegAlign). Alignment analysis of amino acid sequences revealed that, similar to OsCIPK2 and ATCIPK2, HSCIPK2 contains two CIPK-specific functional domains, n...
Embodiment 3
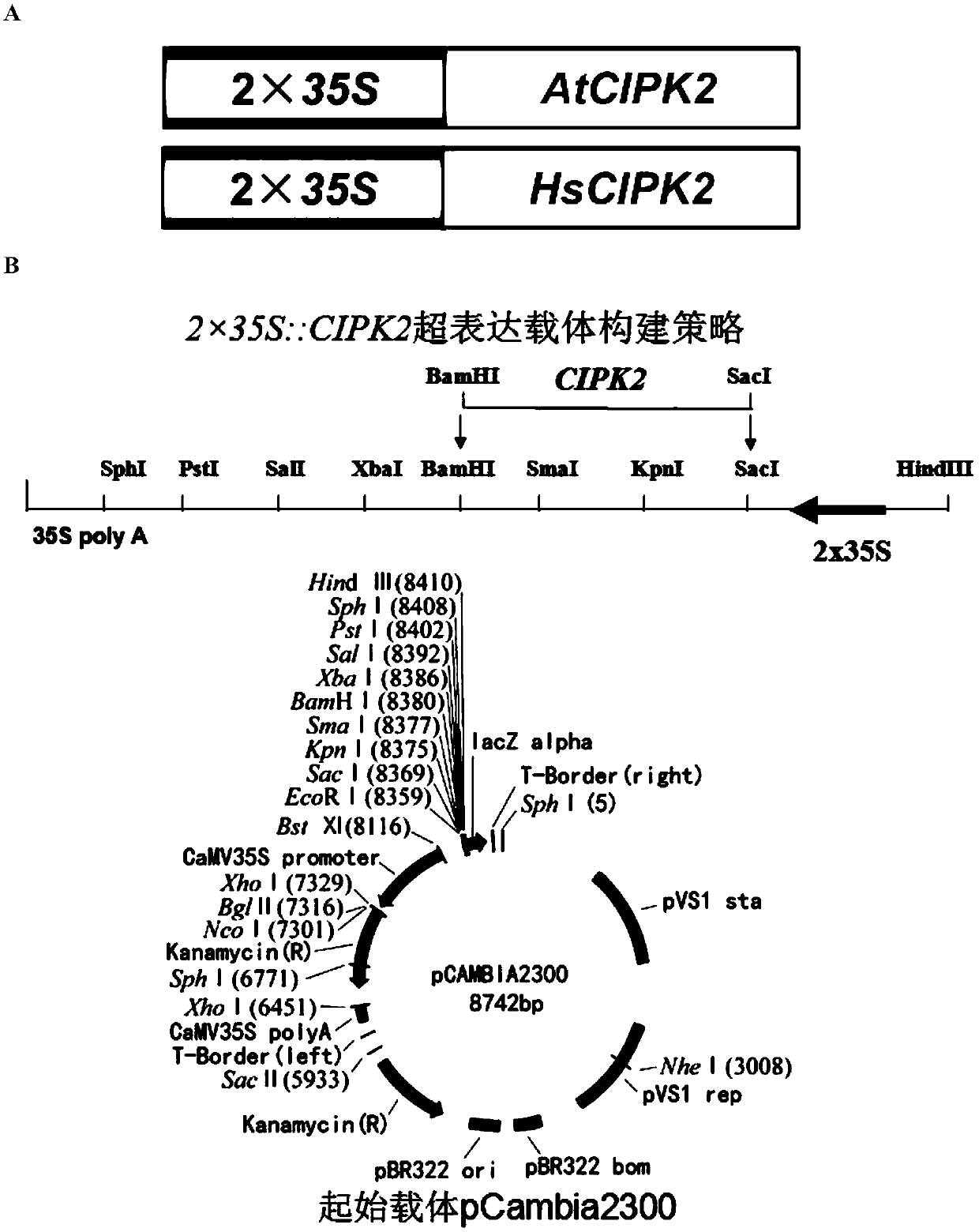
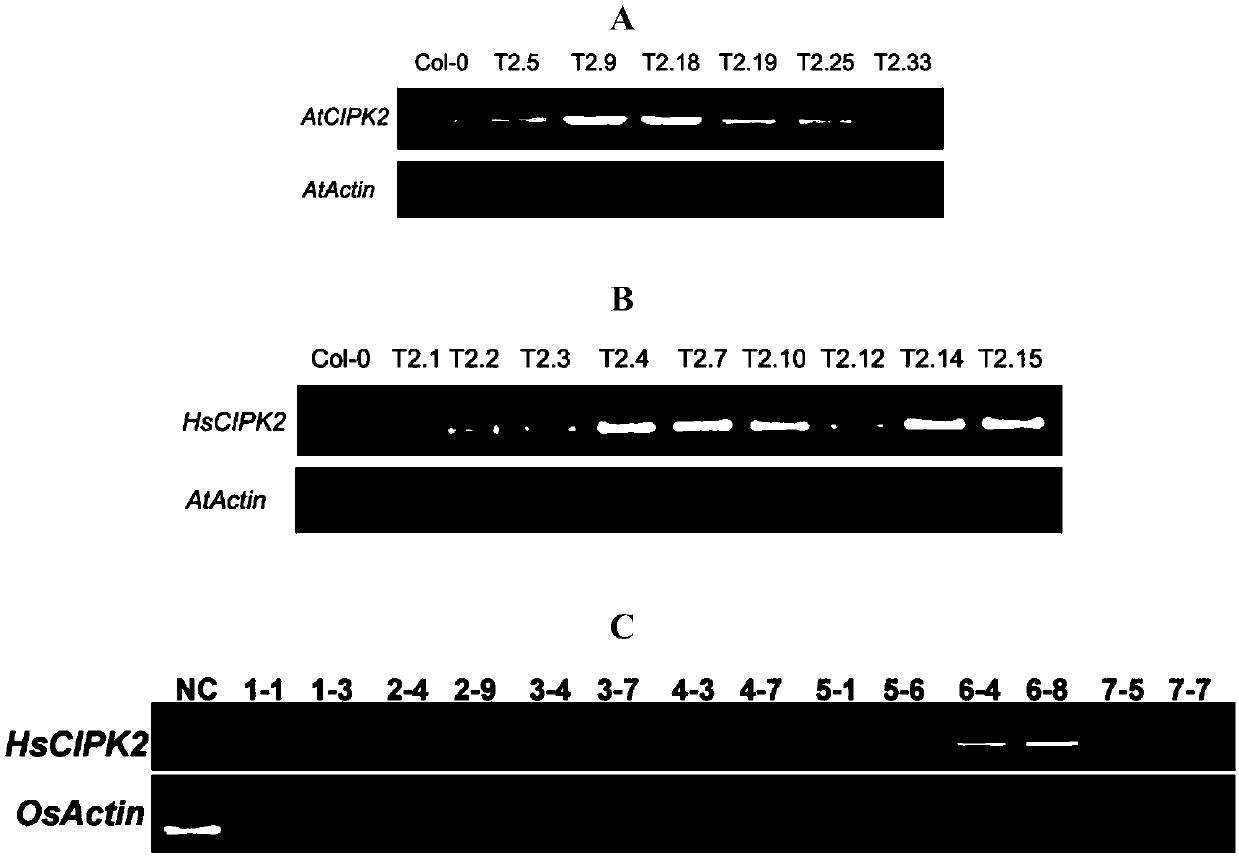
[0077] Embodiment 3, the cultivation of AtCIPK2 and HsCIPK2 gene overexpression transgenic Arabidopsis
[0078] The original recipient material of Arabidopsis thaliana transgene was wild type Arabidopsis (Col-0), and the wild type Arabidopsis Col-0 was introduced into wild type Arabidopsis thaliana Col-0 by inflorescence dipping method mediated by Agrobacterium. Please refer to the published master's degree thesis "Functional Analysis of Arabidopsis Clathrin Light Chain" (Zhejiang Normal University, 2012.4).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com