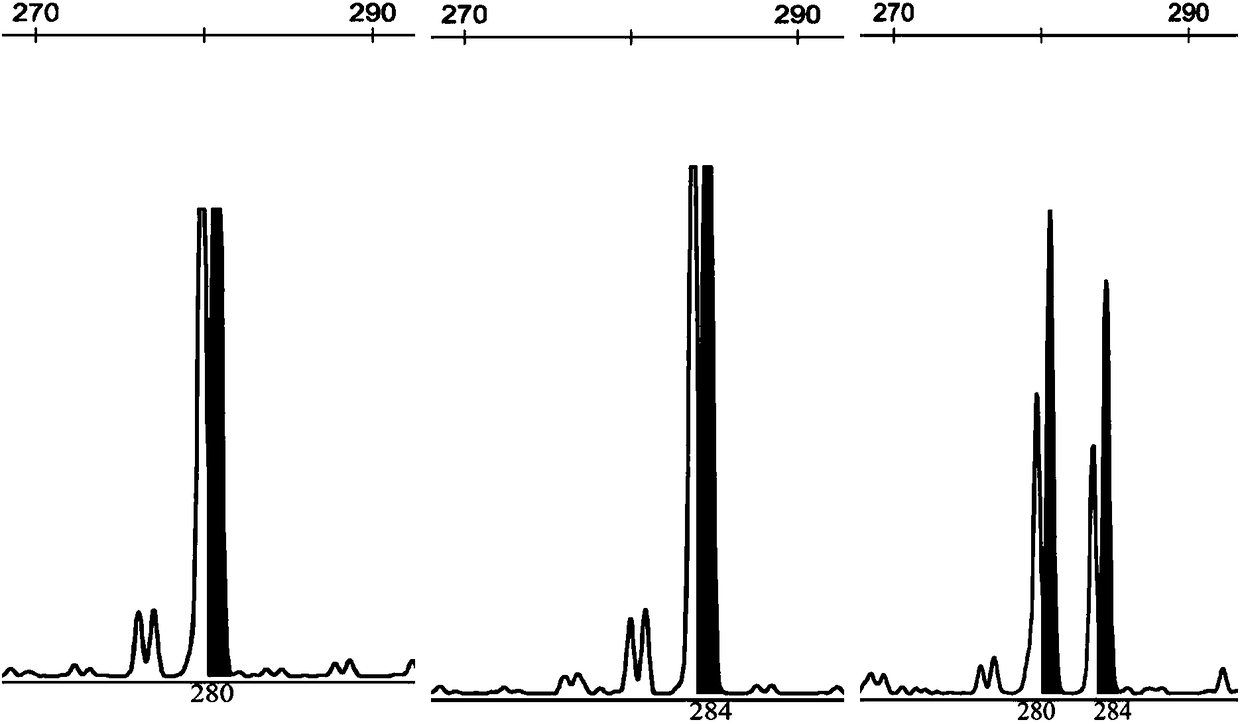
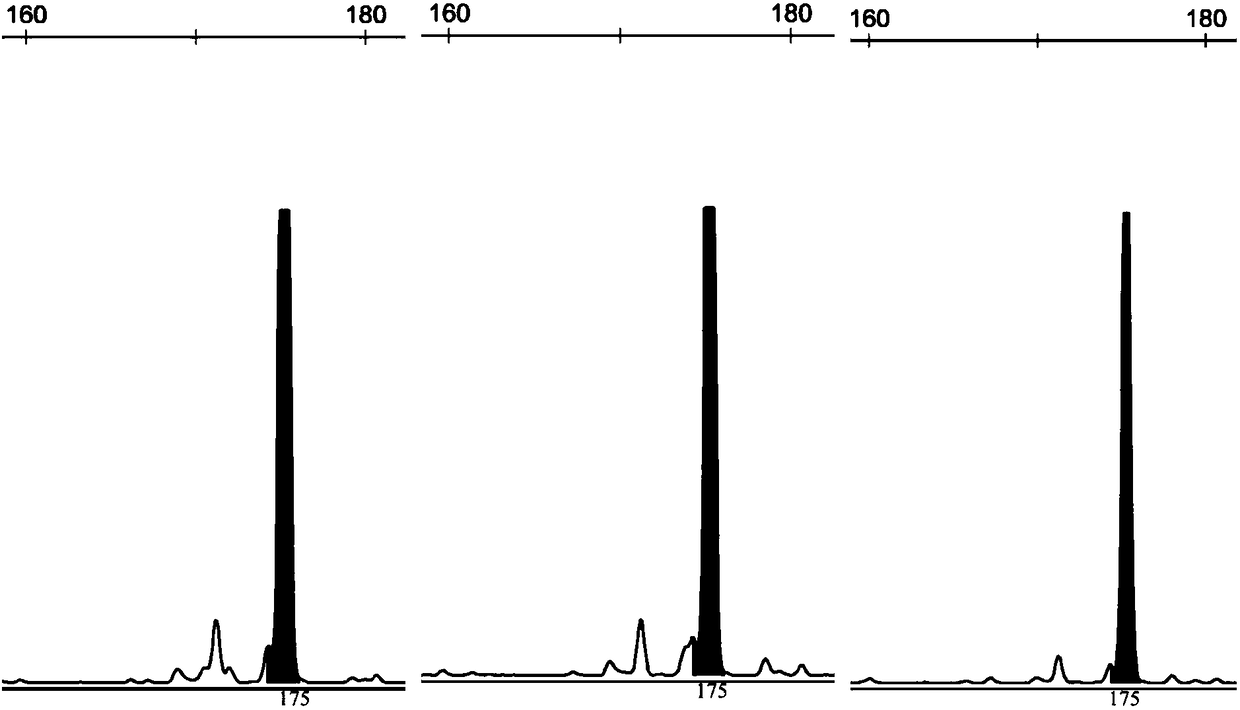
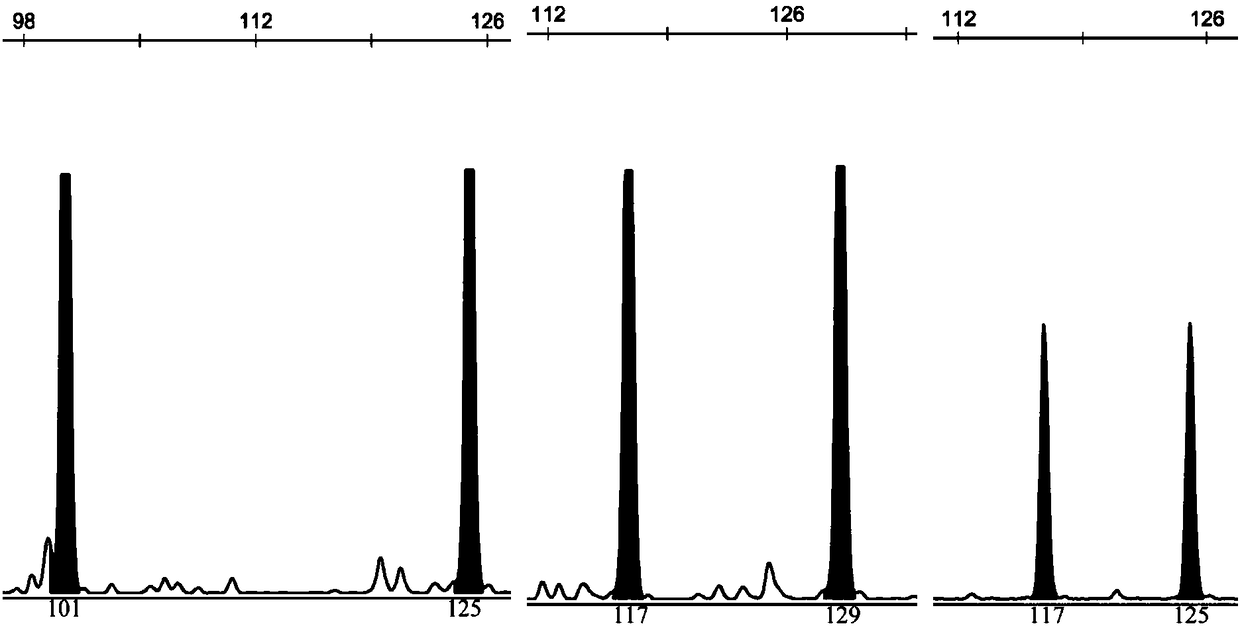
Rhinogobio ventralis Sauvage et Da bry paternity testing method based on micro-satellite fluorescence multiplex PCR
A technology for paternity identification and albacore, which is applied to the determination/inspection of microorganisms, biochemical equipment and methods, recombinant DNA technology, etc., and can solve the problems of paternity identification of albacore and achieve the protection of genetic diversity , improve the accuracy of qualitative, improve the effect of efficiency and speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The steps of polymorphic microsatellite marker screening and multiplex PCR primer combination selection of albacores are as follows:
[0031] 1. Extraction of DNA from individuals of the longfin snout
[0032] Use the Tissue DNA Kit (D3396) kit of Omega Bio-Tek Company to extract the genomic DNA of the sample. The operation steps are as follows: Take 12 wild albacores, cut about 30 mg of fin rays or other tissue samples, wash with double distilled water , cut it thoroughly with small scissors, put it into a 1.5mL centrifuge tube, add 200μL TL Buffer and 25μL OB ProteaseSolution, digest at 55℃ for 1-1.5h, until the tissue is completely digested; centrifuge at 12000rpm for 5min to precipitate insoluble tissue fragments, and aspirate the supernatant Transfer the solution to another 1.5mL centrifuge tube; add 220μL BL Buffer, mix well, and place it in a constant temperature water tank at 70°C for 10min; add 220μL absolute ethanol, mix well, and briefly centrifuge to remove ...
Embodiment 2
[0045] The method for paternity identification of albacores based on microsatellite fluorescence multiplex PCR, the steps are:
[0046] 1. DNA Extraction of Longfin snout individuals
[0047] Collect 6 parental fin ray samples of albacore (3 females and 3 females, reproduced at a ratio of 1:1) and 96 individual samples of the corresponding offspring, and 34 fin ray samples of other breeding parents at the same time, using genomic DNA extraction reagents Genomic DNA was extracted using the kit, and the sample DNA concentration was adjusted to 100ng / μL for storage.
[0048] 2. Microsatellite fluorescent multiplex PCR amplification
[0049] According to the primer sequences and fluorescent labels shown in Table 1, synthesize 15 pairs of albacore microsatellite primers, and label the 5' end of the forward primer of each pair of primers with fluorescent substances, and the genomic DNA in step 1 according to Table 3 The indicated reaction system and annealing temperature were used...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com