Fusion protein hfbi-rgd gene and protein
A technology of fusion protein and gene, applied in the field of protein genetic engineering, to achieve the effect of high yield, good solubility and solving the problem of dispersion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1 Construction of pET-28a-HFBI-RGD:
[0051] With the HFBI nucleotide sequence shown in SEQ ID NO.3 in Trichoderma reesei (Trichoderma reesei) as template, with F1 as forward primer, with R1 as reverse primer, carry out the first round of PCR, obtain the first PCR product; take the first PCR product as the template, use F1 as the forward primer, and use R2 as the reverse primer to carry out the second round of PCR to obtain the first sequence containing the fusion protein HFBI-RGD gene. T4 DNA ligase was ligated, and the fusion protein HFBI-RGD gene was constructed into the PET-28a vector to obtain the PET-28a-HFBI-RGD plasmid. Specific steps are as follows:
[0052] a) Enzyme digestion system
[0053]
[0054] b) Conditions
[0055] i. The target gene was digested for 1 h at 37°C.
[0056] ii. The plasmid was digested for 1 h at 37°C.
[0057] iii. Inactivation at 80°C for 5min after digestion.
[0058] c) Using the HFBI nucleotide sequence shown in SE...
Embodiment 2
[0071] Example 2 Construction of pPIC9k-HFBI-RGD:
[0072] 1) The selection vector is pPIC9k, the restriction sites are NotI and EcoRI, and primers are designed for PCR. The PCR system is shown in Table 3. Primers were designed as F2, R3. Using the plasmid PET-28a-HFBI-RGD constructed in the above Example 1 as a template, PCR can obtain the second sequence containing the fusion protein HFBI-RGD gene with the restriction sites of NotI and EcoRI.
[0073] 2) The above PCR product and the vector pPIC9k were respectively double digested with the restriction enzymes NotI and EcoRI to make the target gene fragment and the vector appear sticky ends. The specific enzyme digestion system and method are as follows:
[0074]
[0075] 3) The target gene fragment with sticky ends and the vector pPIC9k are ligated using T4 DNA ligase, and the ligation system is shown in Table 6 above.
[0076] 4) After transformation and double-enzyme digestion verification, the obtained positive resu...
Embodiment 3
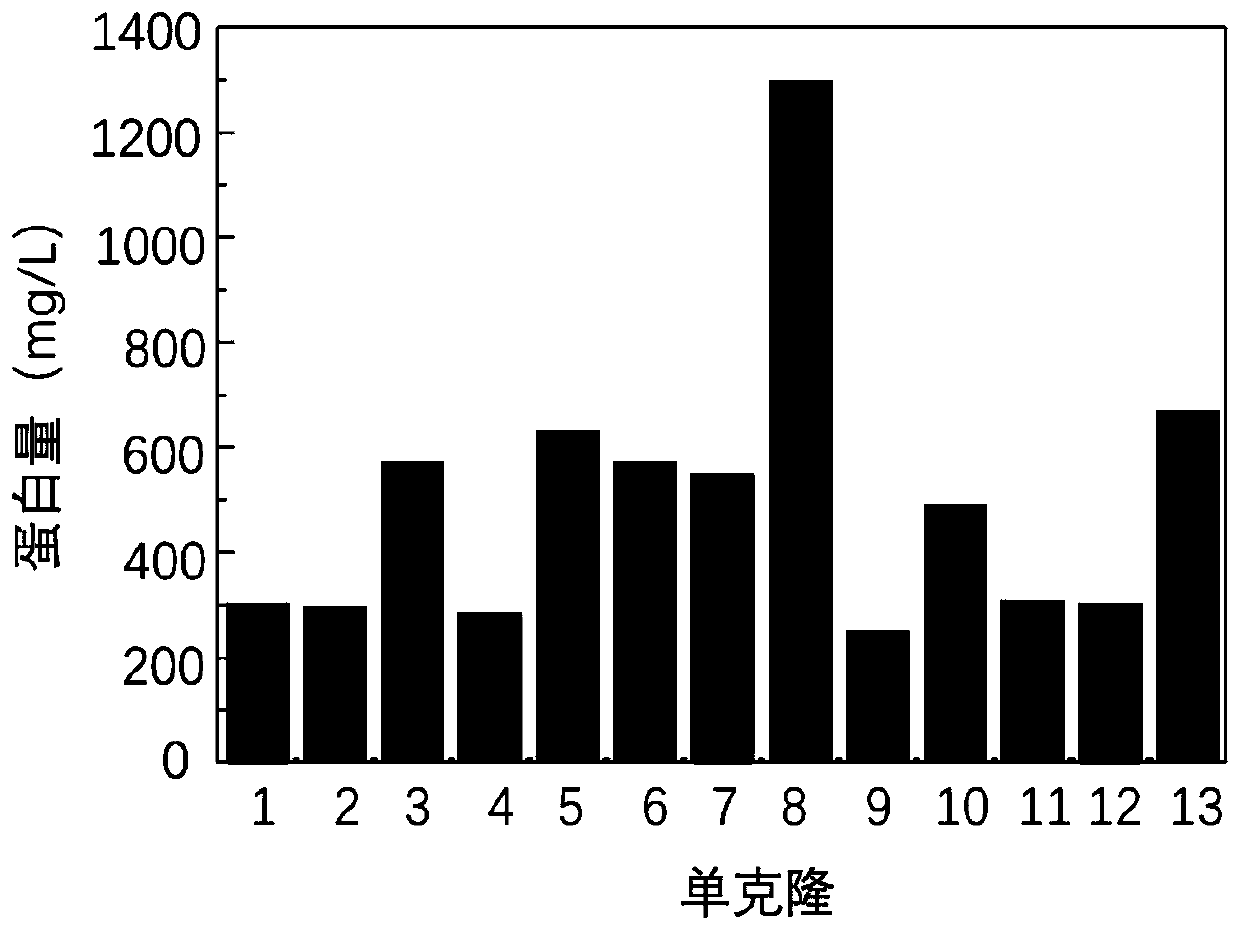
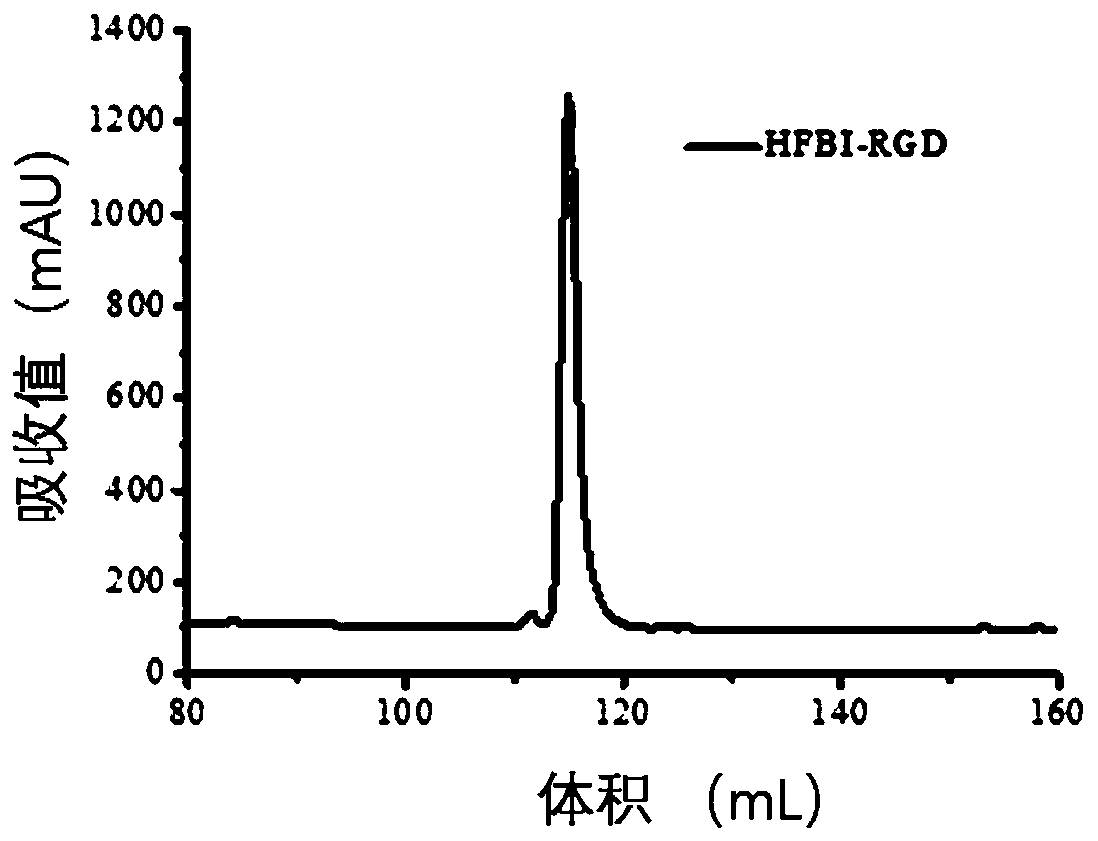
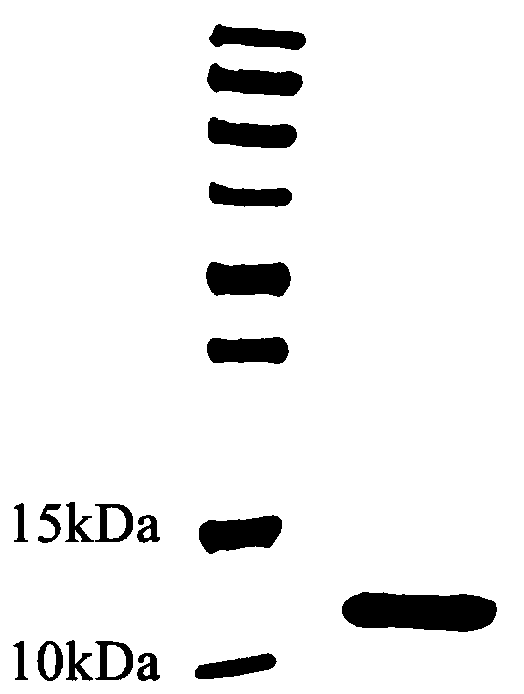
[0077] Example 3 Screening, expression and purification of fusion protein HFBI-RGD positive clones:
[0078] 1) To insert a recombinant expression vector into the yeast genome at a higher copy in yeast cells, it needs to be linearized before being transferred into yeast cells. The enzyme cleavage site analysis function provided in the primer design software Primer 5.0 was used to analyze the cleavage sites of pPIC9k and HFBI-RGD genes, combined with the cleavage site information provided by Ivitrogen in the Pichia pastoris expression system operation manual, and finally The vector was linearized by restriction endonuclease SalI. The reaction system of enzyme cleavage linearization is shown in Table 7. This system was digested in a water bath at 37°C for 2h. Take 5 μL of the product and perform agarose gel electrophoresis to check whether the enzyme digestion is complete, and then the product band is recovered by gel.
[0079] Table 7 Linearization system
[0080]
[008...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com