Application of septin gene shRNA in preparing septin gene activity inhibitor
A technology of sept2-shrna-1 and sept9-shrna-1, which is applied in the field of application of septin gene shRNA in the preparation of septin gene activity inhibitors, can solve problems such as difficult treatment, poor prognosis, difficult resection and treatment, and achieve growth inhibitory effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
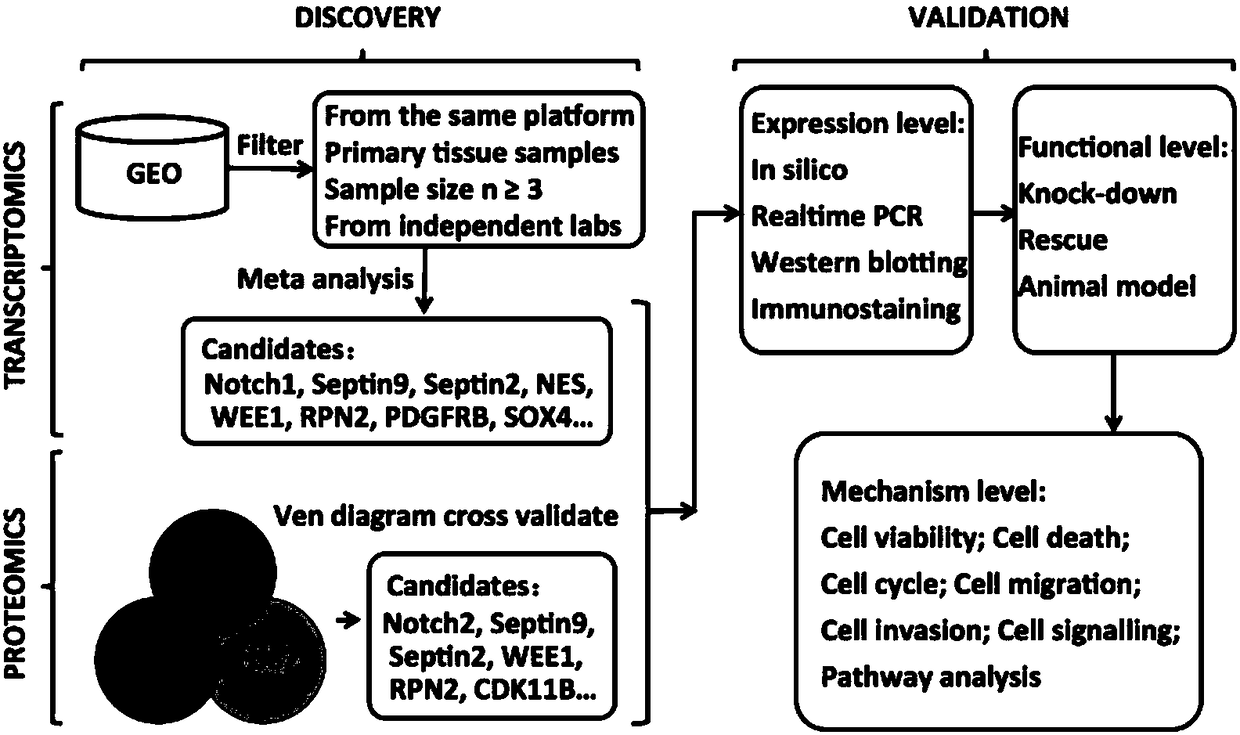
[0025] Example 1: Finding and identifying GBM-related genes through multi-omics analysis
[0026] (1) Experimental method: comprehensive multi-omics analysis
[0027] GBM transcriptomic studies were selected based on the following four criteria: 1) samples were obtained from primary tissues with normal controls; 2) more than three cases and controls were used; 3) experiments were run on the same platform; 4) studies Conducted by an independent panel (see Table 1). Proteomic analysis of three different GBM cell lines was performed simultaneously to represent gene expression at the protein level. For example, the multi-omics analysis workflow flow chart ( figure 1), the data generated by proteomics and transcriptomics are input, and the output is high-quality candidate functional genes, which are arranged according to different statistical criteria. Studies of GBM expression data from the Gene Expression Omnibus (GEO) repository were combined for multiplex analysis. For each...
Embodiment 2
[0032] Example 2: Expression of SEPT9 and SEPT2 in GBM tissues and cell lines
[0033] 1. Experimental method:
[0034] 1. Immunohistochemistry
[0035] (1) Normal brain tissue slices (grade I normal human brain tissue, purchased from Rockville, MD, USA), 3 grades of glioma slices (grade II (astrocytoma), grade III (anaplastic astrocytoma) cell tumor) and grade IV (glioblastoma, GBM, purchased from Rockville, MD, USA) were washed in xylene to remove paraffin and rehydrated by serial dilutions of alcohol.
[0036] (2) Rinse the tissue sections twice with PBS, 5 minutes each time.
[0037] (3) Incubate in 3% hydrogen peroxide for 10 minutes at room temperature to block endogenous peroxidase activity.
[0038] (4) Rinse the tissue sections with PBS for 5 minutes.
[0039] (5) Antigen retrieval. Heat the antigen retrieval buffer (purchased from Sangon Biotechnology) to boiling state with a microwave oven, put the tissue slices into the buffer at 95-100°C for 10 min, twice, co...
Embodiment 3
[0084] Example 3: Inhibition of SEPT9 and SEPT2 expression using shRNA in A172 cells
[0085] 1. Experimental method (fluorescent quantitative PCR):
[0086]
[0087] Total RNA was extracted from GBM cells (ie, A172 cells) and analyzed by real-time quantitative PCR (qRT-PCR) using Trizol reagent (Thermo Fisher Scientific). The amplification reaction was completed by the cfx96 real-time PCR detection system (Bio-Rad Company, Hercules, CA, USA) using the SYBR Green fluorescent quantitative PCR kit according to the operating specifications. For SEPT9 and SEPT2 primers (Table 2), the expression of related genes was calculated using the 2ΔΔCT method. All qRT-PCR experiments were repeated three times, and all data used GAPDH as a control.
[0088] Table 2 SEPT2, SEPT9 qPCR primer sequences
[0089]
[0090] 2. Western blot
[0091] 1) Construction of scramble, SEPT2, SEPT9, SEPT2+SEPT9 plasmids (GFP co-expression): The most commonly used interference plasmid construction m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com