Corn salt-tolerant gene and molecular marker and application thereof
A technology of molecular markers and salt-tolerant genes, which is applied in the fields of application, genetic engineering, plant genetic improvement, etc., to achieve the effect of accelerating genetic improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Genetic Analysis of Maize Salt Tolerance and Mapping and Cloning of Salt Tolerance Genes
[0030] 280 Kyo 724×D9H F 2 For materials, the seeds were sterilized with 1% NaClO for 10 minutes, washed three times with sterilized water, and then planted hydroponically in a corn seedling identification device (disclosed in Chinese patent ZL200920177285.0). The culture conditions are controlled at 26±1°C, 12h light / 12h dark, and the light intensity is 150-180μmol m -2 the s -1 , with a humidity of 70%. After cultivating in an aqueous solution containing 100mMNaCl for 10 days, the root length was used as the salt-tolerant phenotype, and among the 280 materials, 215 materials showed salt tolerance (root length ≥ 5cm), and 65 materials showed salt sensitivity (root length ≥ 5 cm). 2 1:3=0.48, P=0.49), it can be seen that the salt tolerance of maize root length is controlled by a dominant single gene.
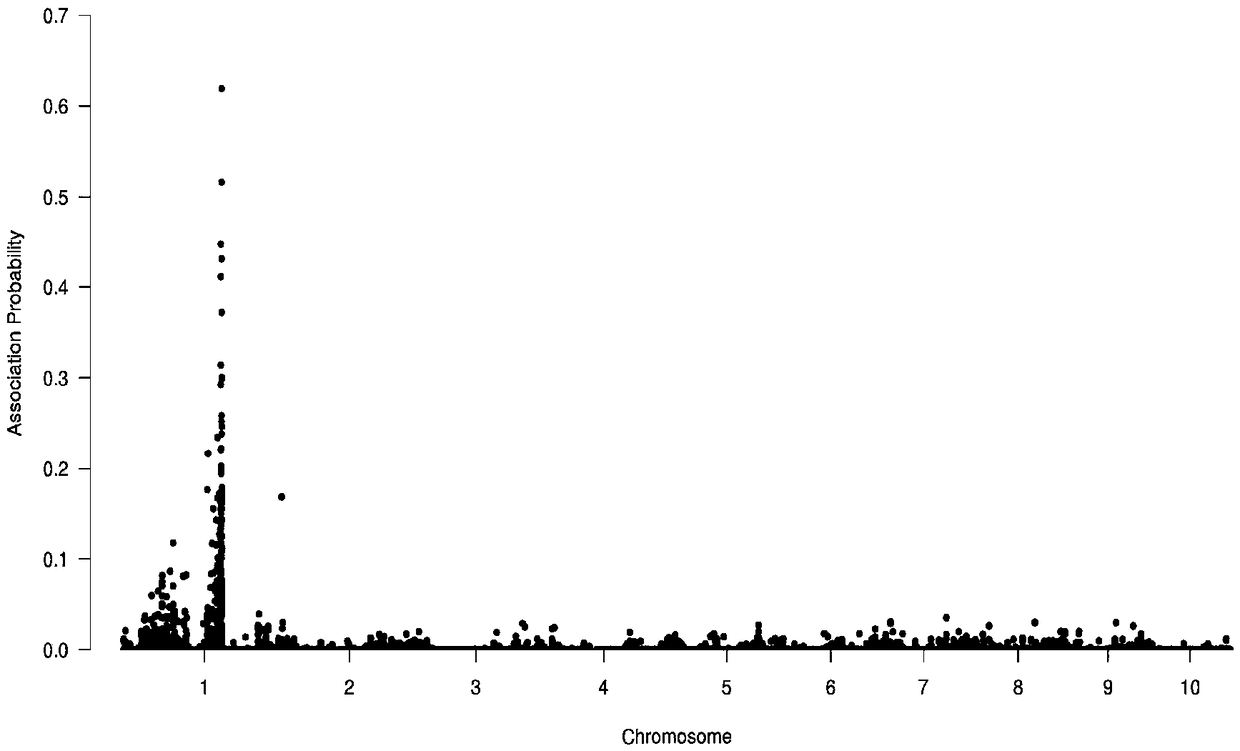
[0031] Preliminary mapping of salt tolerance genes using BSR-seq ...
Embodiment 2
[0045] Example 2 Development and Application of INDEL Molecular Markers Related to Maize Salt Tolerance
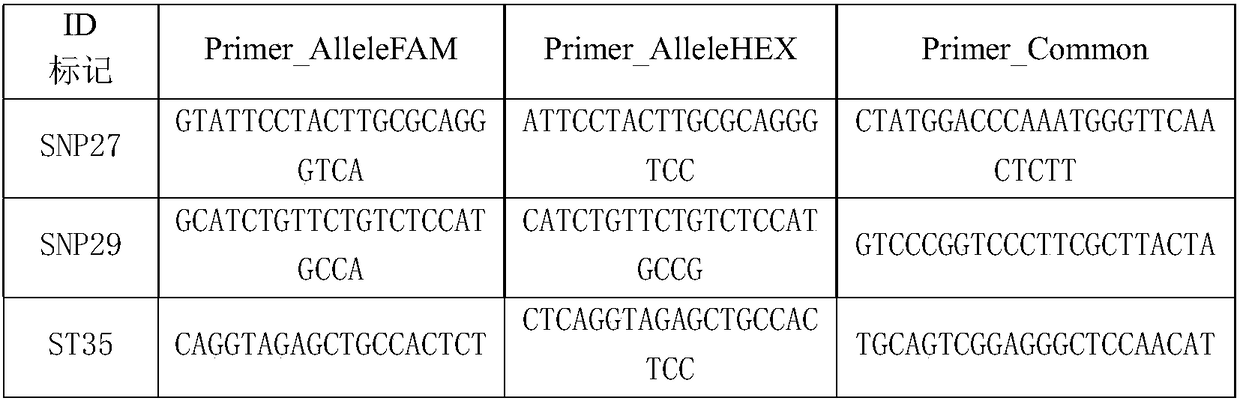
[0046] ZmST1 has a 4 bp base insertion / deletion difference between the salt-tolerant maize inbred line Jing 724 and the salt-sensitive inbred line D9H, and functional molecular markers were developed based on the sequence differences. The functional molecular marker ST35 functional site developed is TCTT / -, its KASP primer information is shown in Table 1. After culturing 50 parts of corn DH series materials in an aqueous solution containing 100 mM NaCl for 10 days, the root length was measured with a ruler, and leaf DNA was extracted with a plant DNA extraction kit from TianGen Company. Using this functional molecular marker, according to the standard experimental procedures of Laboratory of the Government Chemist (LGC) company, the KASP method was used to detect the InDel site, and it was found that the salt tolerance obtained by applying the functional molecular marker ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com