Method and kit for detecting m<7>G (N7-methylguanosine) modification in RNA with single base resolution in whole transcriptome range
A transcriptome, single-base technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as increased harm and inapplicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
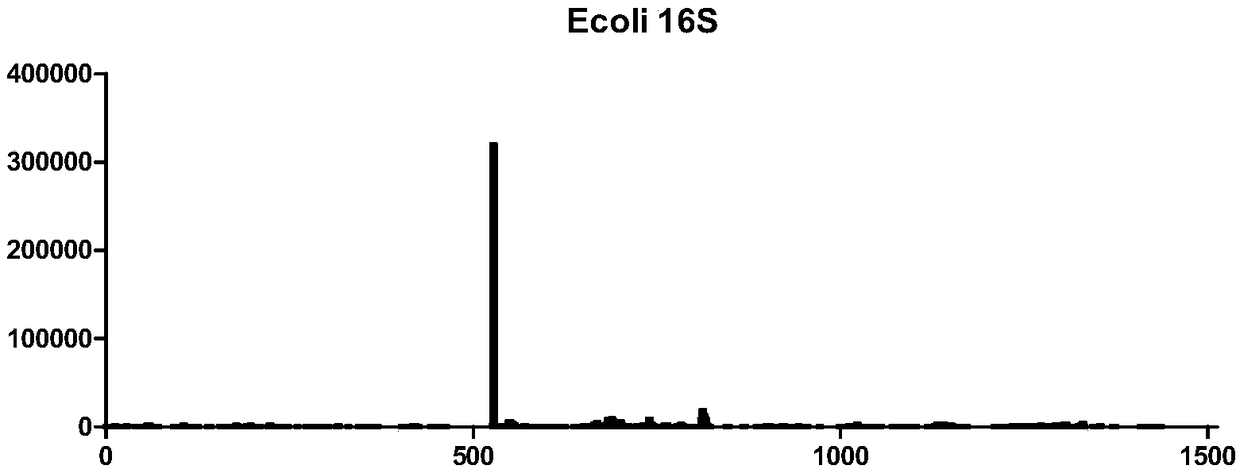
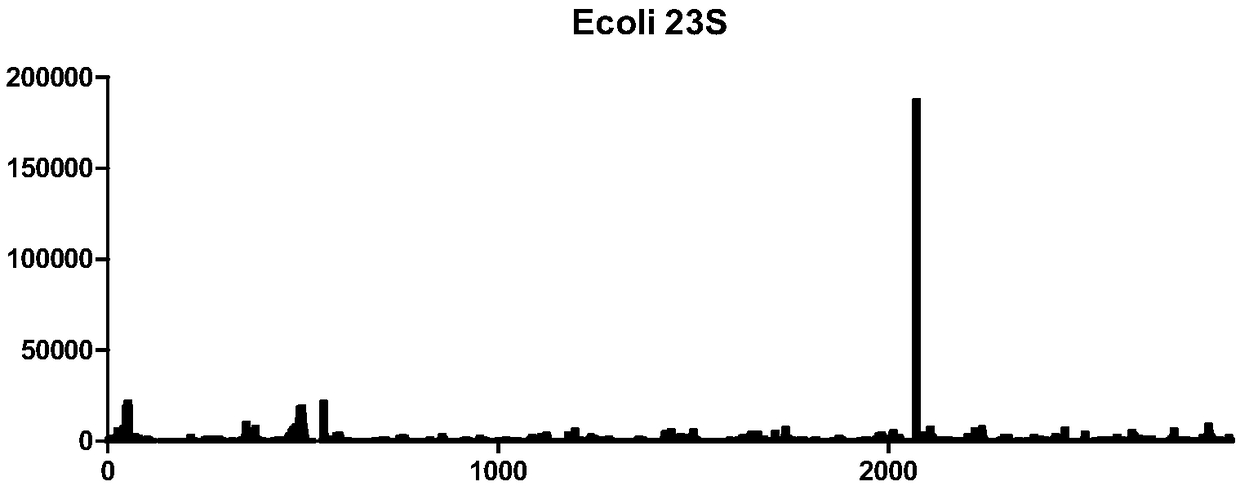
[0031] Example 1 Detection of RNA7mG modification in E. coli
[0032] 1. RNA acquisition
[0033] (1) Centrifuge the 100 mL of Escherichia coli cultured overnight, at 4000 rpm, 4°C for 5 minutes, remove the supernatant, and retain the bacterial pellet;
[0034] (2) Put the prepared mortar and pestle on a tray filled with water, pour a little alcohol and ignite it, take out the mortar and pestle after the alcohol is burned and cooled;
[0035] (3) Introduce liquid nitrogen into the mortar, use a syringe to suck the bacterial pellet and drop the precipitate into the liquid nitrogen drop by drop. After the liquid nitrogen is converted, use a pestle to grind and grind into powder, and then add liquid nitrogen. After the liquid nitrogen disappears, immediately use a pestle to grind, repeat this three times, and finally add Trizol Reagent immediately (add more points), and then Trizol Reagent will condense into a block;
[0036] (4) After Trizol Reagent becomes liquid, pipette and mix well, ...
Embodiment 2
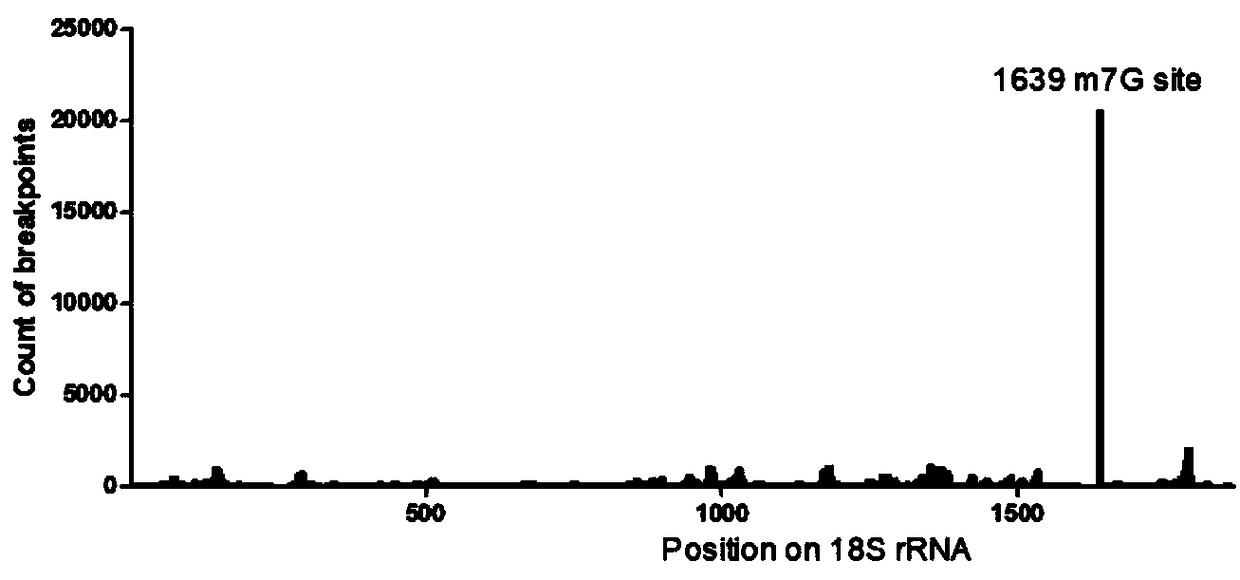
[0102] Example 2 Detection of RNA7mG modification in HEK293T cell line
[0103] 1. RNA acquisition
[0104] (1) Digest the cells cultured in a 10cm diameter cell culture dish with trypsin, collect them in a 15mL centrifuge tube, centrifuge at 1000rpm for 5 minutes, remove the supernatant, and save the cell pellet;
[0105] (2) Resuspend the cells in 10 mL PBS buffer, centrifuge for 5 minutes, remove the supernatant, and save the cell pellet;
[0106] (3) Add 2 mL of LTrizol Reagent to the cell pellet, pipette to mix, and then divide 1 mL per tube into 1.5 mL EP tubes, then shake and mix with a shaker, and place on ice for 5 minutes to make the cells full Cracking
[0107] (4) Add 1 / 5 chloroform according to the volume of TRI Reagent, mix vigorously with a shaker for 15 seconds, and place on ice for 10 minutes;
[0108] (5) Centrifuge at 12,000rpm for 15 minutes at 4°C, transfer the upper aqueous phase to a new EP tube, taking care not to absorb the middle organic solvent layer. Add an ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com