Detection method for low-frequency mutation of circulating tumor DNA
A detection method and low-frequency mutation technology, applied in the field of high-throughput sequencing, can solve the problems of increasing the proportion of dominant molecular sequences, the number of molecular labels is small, and the frequency is not exactly the same, so as to reduce the data repetition rate, avoid consecutive bases, and save The effect of data volume
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] 10ml of whole blood was collected from 5 patients with lung cancer, and the collection tubes were Streck Cell-Free DNA BCT® BloodCollection Tubes (10ml), stored and transported at room temperature for no more than 72 hours. Use two-step method to separate plasma. First, centrifuge the whole blood at 1600g for 10min at room temperature, take out about 4.5ml of supernatant and put it in a 15ml centrifuge tube, then centrifuge at 16000g for 10min at 4℃, take out about 4ml of supernatant, and put it in a 1.5ml centrifuge tube. Store frozen at -80°C. Plasma cfDNA was extracted using QIAamp @ ciculiatiq NucleicAcid KIT, and finally eluted with 50ul EB buffer. The sample name and extracted cfDNA concentration and quality information are shown in Table 4:
[0031]
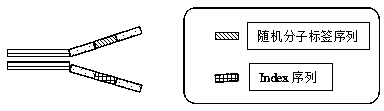
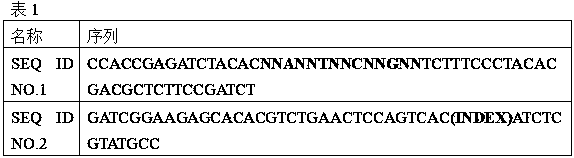
[0032] Synthesize adapters with random labels as shown in the above table 1, wherein, N represents any one of the four bases of ATCG, and every two random bases are separated by an A or T or C or G base, A total...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com