Laying duck circRNA circ_13034 and detection reagent, method and application thereof
A detection kit and a technology for laying ducks are applied in the field of genetic engineering to achieve the effect of improving the fecundity of laying ducks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Laying duck circular RNA circ_13034 gene
[0035] 1. Laying duck circular RNA circ_13034
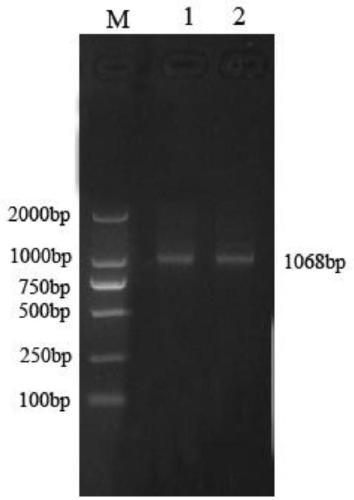
[0036] The cDNA sequence corresponding to the laying duck circular RNA circ_13034 described in this example is shown in SEQ ID NO: 1. The laying duck circular RNA circ_13034 of the present invention is based on the whole transcriptome sequencing result of the laying duck follicle tissue completed earlier by the applicant. A new circular RNA molecule was obtained through extensive bioinformatics analysis and screening of the sequencing results of two groups of follicular tissue samples at different developmental stages. The applicant named it egg-duck circular RNA circ_13034, and the length of the cDNA sequence corresponding to the egg-duck circular RNA circ_13034 was 1068 bp.
[0037] 2. Preparation of the full-length sequence fragment of circular RNA circ_13034 gene of laying duck
[0038] 1. Primer design and synthesis
[0039] According to the full-length fragment ...
Embodiment 2
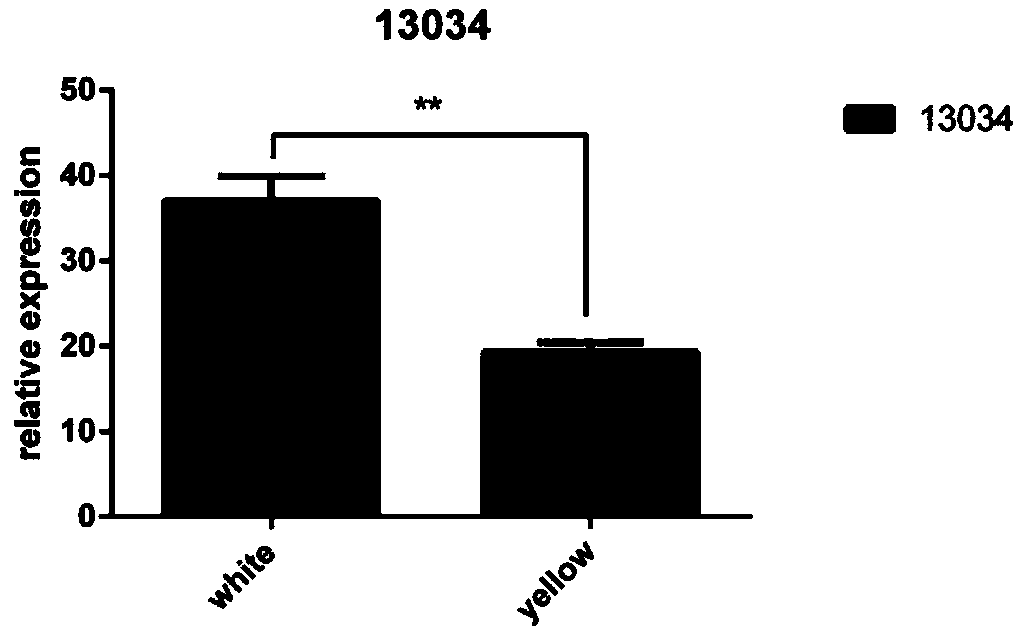
[0084] Example 2 Fluorescent quantitative PCR detection primer pair and its detection kit and method for laying duck circular RNA circ_13034 gene
[0085] In this example, a pair of fluorescent quantitative PCR detection primers was designed to identify and identify the objective existence of the laying duck circular RNA circ_13034 described in Example 1 in the egg follicle granulosa cells of the laying duck. The identification method of this embodiment includes the following steps:
[0086] 1. Sample collection
[0087] With the method in embodiment 1.
[0088] 2. Extraction of total RNA from samples
[0089] Same as the RNA extraction method in Example 1.
[0090] 3. RNA sample quality detection and analysis
[0091] With the method in embodiment 1.
[0092] 4. Digestion of circular RNA (RNase R)
[0093] With the method in embodiment 1.
[0094] 5. Reverse transcription
[0095] With the method in embodiment 1.
[0096] 6. Fluorescent quantitative PCR reaction
[...
Embodiment 3
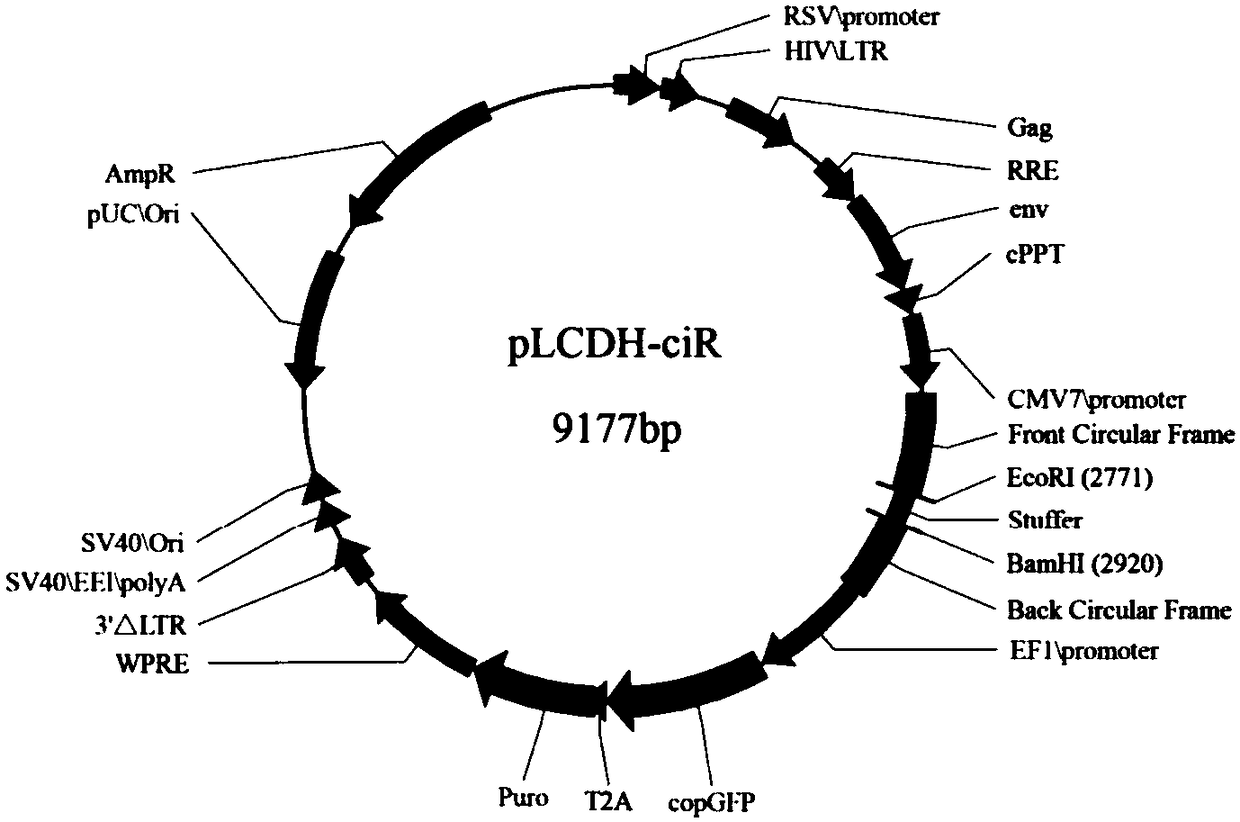
[0104] Example 3 Construction of egg duck circular RNA circ_13034 gene overexpression vector
[0105] This embodiment provides an overexpression vector of the egg duck circular RNA circ_13034 gene, and the vector includes the cDNA sequence corresponding to the egg duck circular RNA circ_13034. Vector construction specifically includes the following steps:
[0106] (1) Double digestion treatment of pLCDH-ciR vector
[0107] The schematic diagram of the structure of the pLCDH-ciR vector is shown in image 3 As shown, the vector contains restriction endonucleases EcoRI and BamHI cutting sites. The above carrier was double-digested with two restriction enzymes EcoRI and BamHI. The enzyme digestion reaction system was as follows, and the total volume of the system was 50 μL:
[0108] table 5
[0109]
[0110] The reaction conditions are: enzyme digestion at 37°C for 6 hours, followed by purification and recovery of the enzyme digestion products.
[0111] (2) Double enzyme d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com