Mutated immunoglobulin-binding polypeptides
A technology that combines peptides and staphylococcal proteins, applied in the direction of immunoglobulin, carrier-bound/immobilized peptides, peptides, etc., can solve the problems of difficult to achieve cleaning, unacceptable loss of ability, etc., and achieve the effect of highly selective binding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
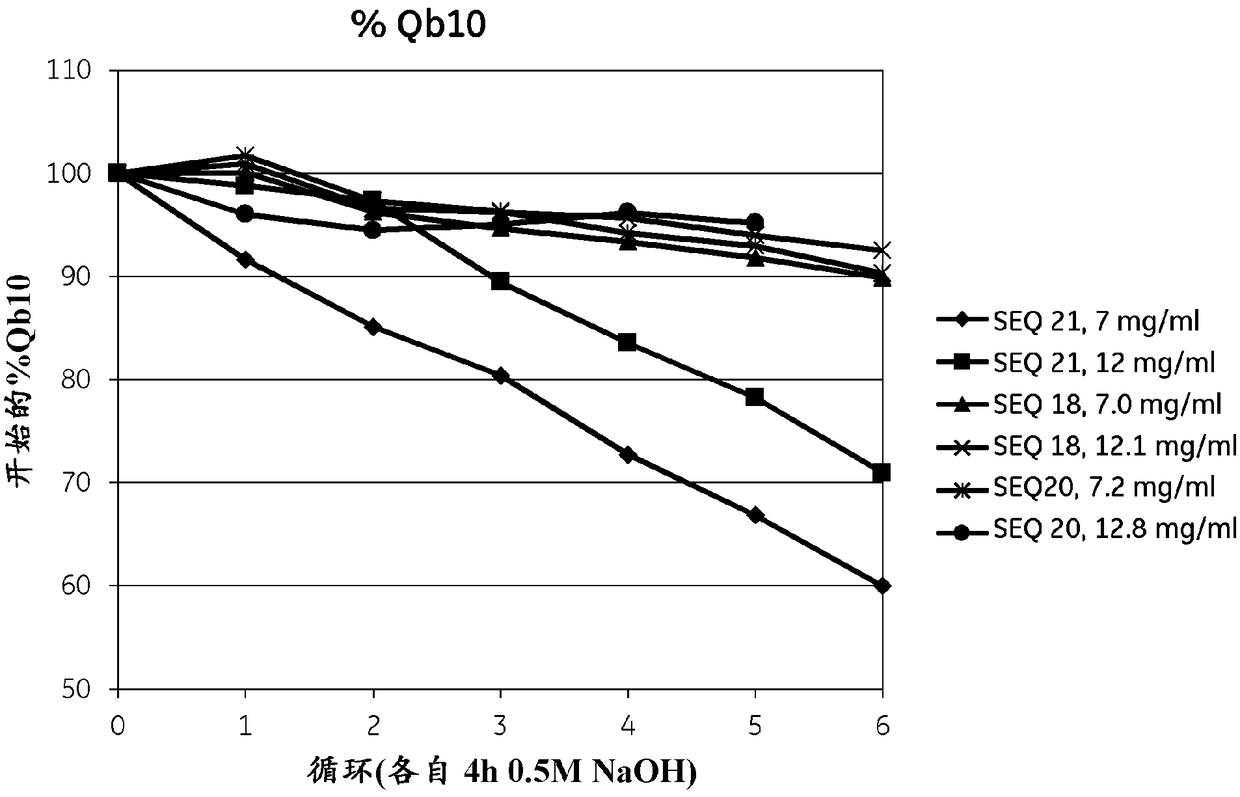
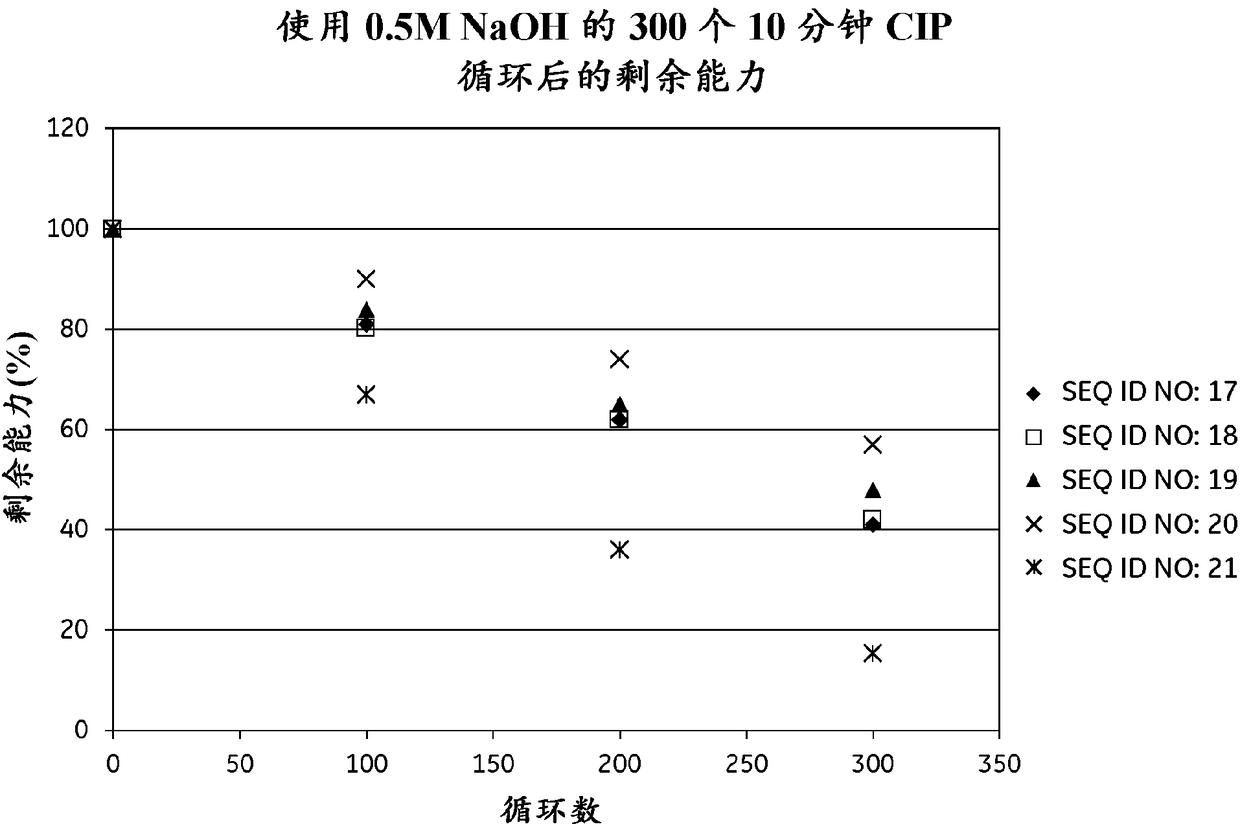
[0293] The purified monomeric ligands listed in Table 1 (for SEQ ID NOs 8-16, 23-28 and 36-48, also containing the AQGT leader sequence at the N-terminus and the cysteine at the C-terminus), Using GE Healthcare's Amine Coupling Kit (for carbodiimide coupling of amines on carboxymethyl groups on a chip) sufficient to give approx. Amounts of signal intensity ranging from 200-1500 RU were immobilized on Biacore CM5 sensor chips (GE Healthcare, Sweden). To follow the IgG binding capacity of the immobilized surface, 1 mg / ml human polyclonal IgG (Gammanorm) was flowed over the chip and the signal intensity (proportional to the amount bound) was recorded. The surface was then cleaned in place (CIP), ie rinsed with 500 mM NaOH for 10 minutes at room temperature (22 + / - 2°C). 96-100 cycles were repeated and the immobilized ligand alkali stability was followed after each cycle by remaining IgG binding capacity (signal intensity). The results are shown in Table 1 and show that at lea...
Embodiment 2
[0300]Purified dimer, tetramer and hexameric ligands listed in Table 2 using GE Healthcare's Amine Coupling Kit (for carbodiimide coupling of amines on carboxymethyl groups on the chip) , was immobilized on a Biacore CM5 sensor chip (GE Healthcare, Sweden) in an amount sufficient to give a signal intensity of approximately 200-1500 RU in a Biacore instrument (GE Healthcare, Sweden). To follow the IgG binding capacity of the immobilized surface, 1 mg / ml human polyclonal IgG (Gammanorm) was flowed over the chip and the signal intensity (proportional to the amount bound) was recorded. The surface was then cleaned in place (CIP), ie rinsed with 500 mM NaOH for 10 minutes at room temperature (22 + / - 2°C). 300 cycles were repeated and the immobilized ligand alkali stability was followed after each cycle by remaining IgG binding capacity (signal intensity). The results are shown in Table 2 and figure 2 and showed that at least the ligands Zvar(Q9A,N11E,N43A)4, Zvar(Q9A,N11E,N28A,N...
Embodiment 3
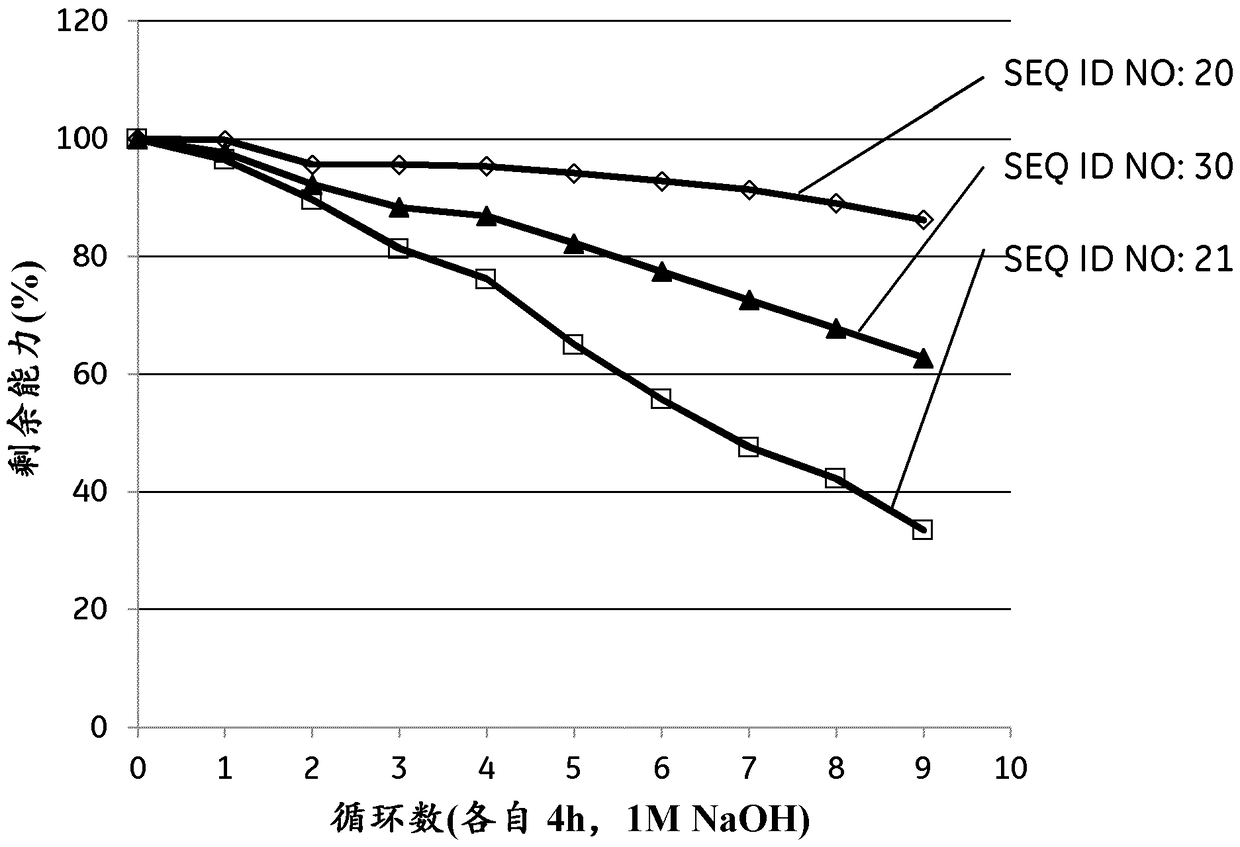
[0305] Example 2 was repeated with 100 CIP cycles of 3 ligands using 1 M NaOH instead of 500 mM in Example 2. The results are shown in Table 3 and show that all 3 ligands also have improved alkaline stability in 1M NaOH compared to the parental structure Zvar4 used as reference.
[0306] Table 3. Tetrameric ligands evaluated with Biacore (1M NaOH).
[0307] Ligand
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com