A functional marker for detecting a broad-spectrum rice blast resistance gene PigmR and an application thereof
A rice blast resistance gene and function technology, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of easy loss of target genes, large genetic distance, etc. Long cycle and stable effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: PigmR gene sequence analysis and design of functional markers
[0044] Currently, the molecular markers for identifying the PigmR gene are linkage markers. The present invention designs functional markers according to the functional sequence characteristics of the PigmR gene, so as to improve the identification effect and breeding application of the PigmR gene.
[0045] In Gumei No. 4 (from the Agricultural Germplasm Bank of Jiangsu Province), the Pigm gene cluster contains 13 R genes (R1-R13), among which R4, R6 and R8 can form complete transcripts. R6 is a disease resistance gene, which is a functional Pigm gene, called PigmR (sequence shown in SEQ ID NO.21), and there are no alleles in varieties or germplasms such as Nipponbare and 9311; R4 and PigmR genes are in protein There is a difference of 4 amino acids at the level, which has no function, and is called Pigm-R4 (sequence shown in SEQ ID NO.22); R8 is a susceptible gene, called PigmS (sequence shown i...
Embodiment 2
[0049] Example 2 Detection and optimization of functional markers
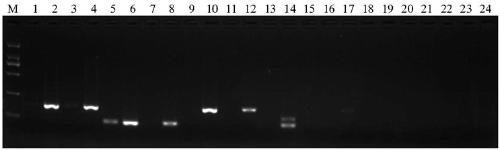
[0050] Genomic DNA from seedling leaves of Gumei 4, Nipponbare and Huanghuazhan was extracted by CTAB method (Murray M G, et al., Nucleic Acids Research, 1980, 8(19):4321-4326). Using the genomes of Gumei No. 4, Nipponbare and Huang Huazhan as templates, PCR screening was performed on the molecular markers for detecting the PigmR gene in Table 1. The PCR reaction system is 2 μL of genomic DNA, 2 μL of 10×PCR buffer, MgCl 2 (5mmol / L) 2μL, dNTP (2mmol / L) 2μL, upstream primer (2μmol / L) 2μL, downstream primer (2μmol / L) 2μL, Taq enzyme 0.2μL, ddH 2 O7.8 μL. The amplification conditions were 94°C for 5min; 35 cycles of 94°C for 30s, 55°C for 30s, and 72°C for 30s; 72°C for 10min to end the reaction. PCR reactions were performed in an Eppendorf Mastercycle thermal cycler. Amplified products were separated by agarose gel electrophoresis and photographed in a gel imager, such as figure 1 shown. The results showed...
Embodiment 3
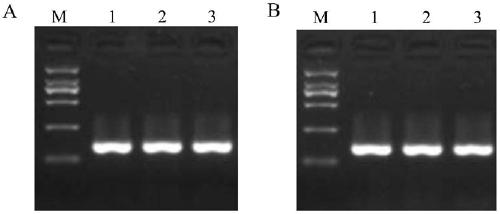
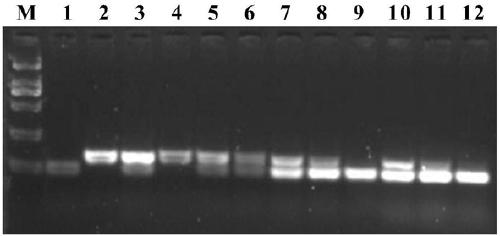
[0052] Example 3: Comparison of GMRA markers and Indel587 markers for detection of rice varieties
[0053]Indel587 is currently a conventional linkage marker for identifying the PigmR gene. The pigmR gene functional marker GMRA marker and the Indel587 marker developed by the inventors were used to detect and compare rice varieties. For the specific detection method, refer to the PCR detection method in Example 2 for detection. The PCR reaction system is 2 μL of rice plant genomic DNA, 2 μL of 10×PCR buffer, MgCl 2 2 μL, dNTP 2 μL, GMR-3F 4 μL, GMR-3R 4 μL, Actin1-1F 1 μL, Actin1-1R 1 μL, Taq enzyme 0.2 μL, ddH 2 O 1.8 μL. Amplification conditions were 94°C for 5 minutes; 94°C for 30s, 55°C for 30s, 72°C for 30s, 35 cycles; 72°C for 10 minutes, and the reaction was terminated. For the primer concentration, refer to Example 2 for the optimal concentration. The detection materials include indica rice plum No. 4, Nipponbare, Huang Huazhan, IR36, Yuzhenxiang, Nanjing No. 16, Kas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com