Application of transcriptomics for co-expressing cyclic circadian rhythm to discovery of drug action mechanism
A technology of drug action and circadian rhythm, which can be used in chemical property prediction, chemical process analysis/design, molecular entity identification, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
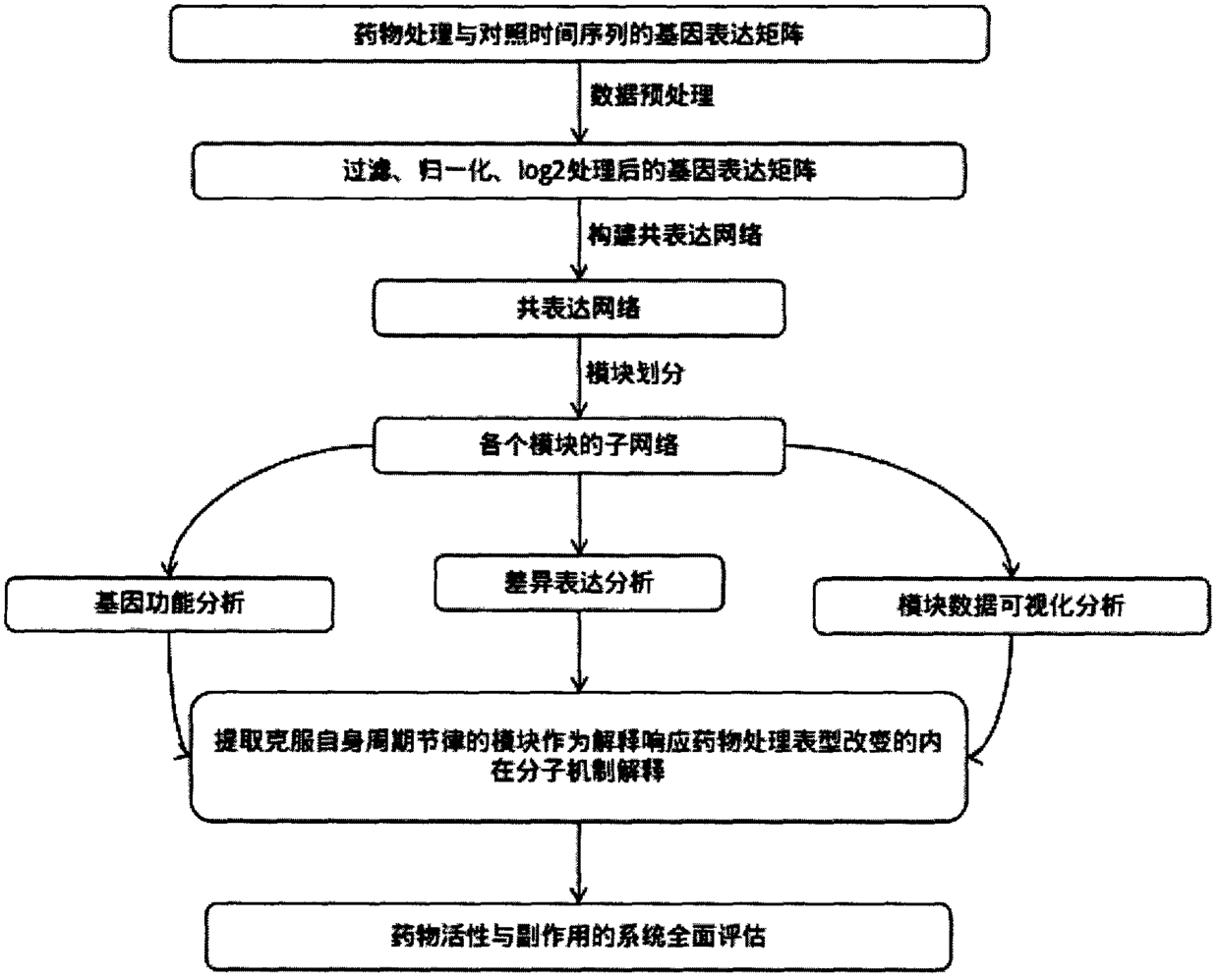
[0041] Example 1: Discovery process route for drug action mechanism of co-expression cycle circadian rhythm transcriptomics
[0042] The research platform for the mechanism of action between drugs and organisms with drug-biological phenotypes is: use mRNA expression data to construct mRNA expression networks and divide them into modules, and then use hypergeometric distribution to enrich the functions of GO functions and KEGG pathways for each module Analysis, and then extract the module that overcomes its own cycle rhythm as the main explanation of the internal molecular mechanism of the corresponding drug treatment-related biological phenotype changes, and finally integrate the phenotypic changes and functional analysis after drug treatment and use data visualization to carry out the physiological effects of drugs on the corresponding biological phenotypes The molecular mechanism of the function is fully explained and evaluated; this process route can also be used to study th...
Embodiment 2
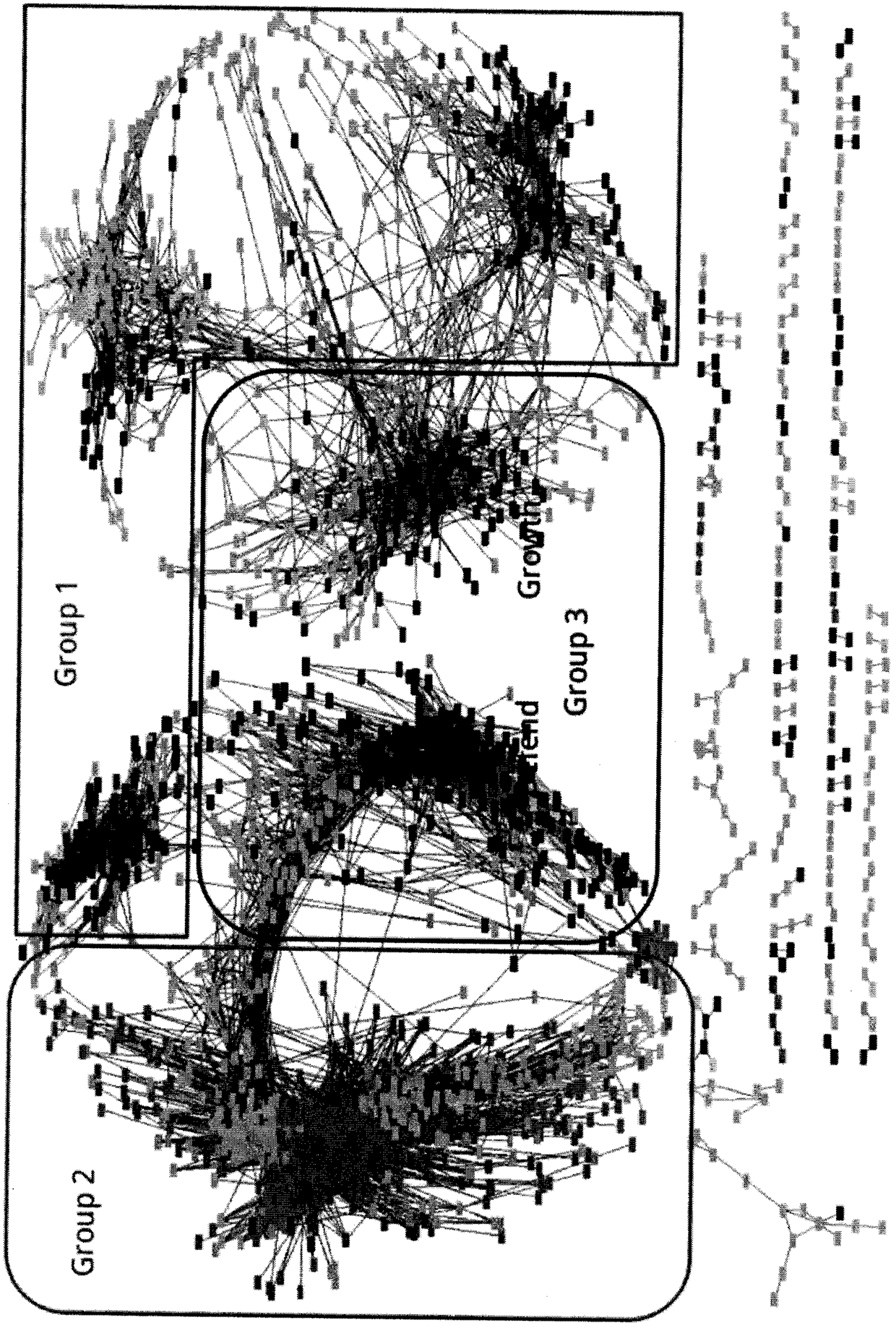
[0043] Example 2: A method for discovering the mechanism of action of a pesticide, taking the mechanism of coronatin as an example of inhibiting growth and promoting defense
[0044] In this example, the present invention is applied to the mRNA expression data of the two-week-old Arabidopsis seedlings treated with coronatin to cause the growth of the seedlings to stop, and the resistance to pathogenic bacteria and insect pests to be enhanced. The time span is 0h, 0.25h, 0.5h, 1h, 1.5h, 2h, 2.5h, 3h, 4h, 5h, 6h, 7h, 8h, 10h, 12h, 14h, 16h, 18h, 20h, 22h, 24h, h is the abbreviation of time hours. According to the drug treatment, the samples were divided into the control group, the control group was represented by mock and the experimental group, and the experimental group was represented by COR, and the common gene co-expression network of the control group and the treatment group was constructed by using the WGCNA method.
[0045] See the attached manual for the specific proces...
Embodiment 3
[0058] Example 3: Transcriptomics of Co-expression Periodic Circadian Rhythm to Explore the Mechanism of Drug Action and Its Process Route and Operation Steps Used in the Field of Medicine to Discover the Mechanism of Drug Action
[0059] Using the experimental medical animal and human mRNA expression data to construct the mRNA expression network and divide it into modules, then use the hypergeometric distribution to analyze the GO function and KEGG pathway function enrichment of each module, and then extract the module that overcomes its own cycle rhythm as the corresponding medical treatment The main explanation of the internal molecular mechanism of the phenotypic changes of related experimental medicine animals and humans. Finally, the phenotypic changes and functional analysis after comprehensive medical treatment and the use of data visualization are used to fully explain the molecular mechanisms of the corresponding experimental medicine animals and human physiological fu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com