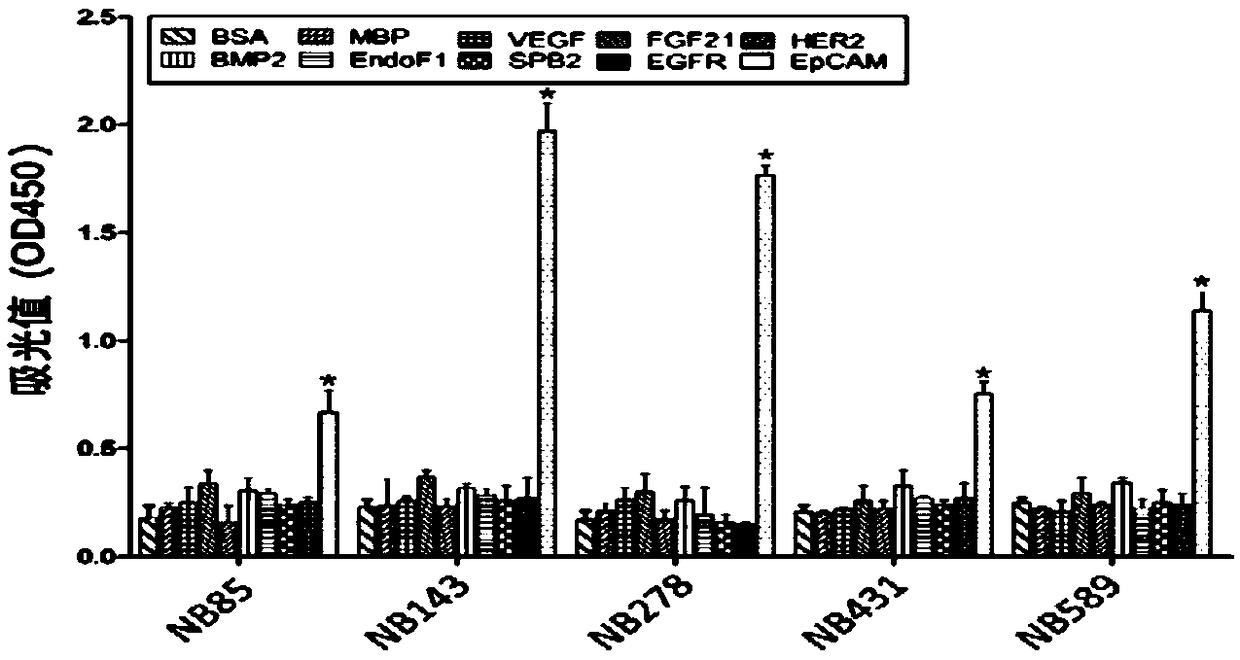
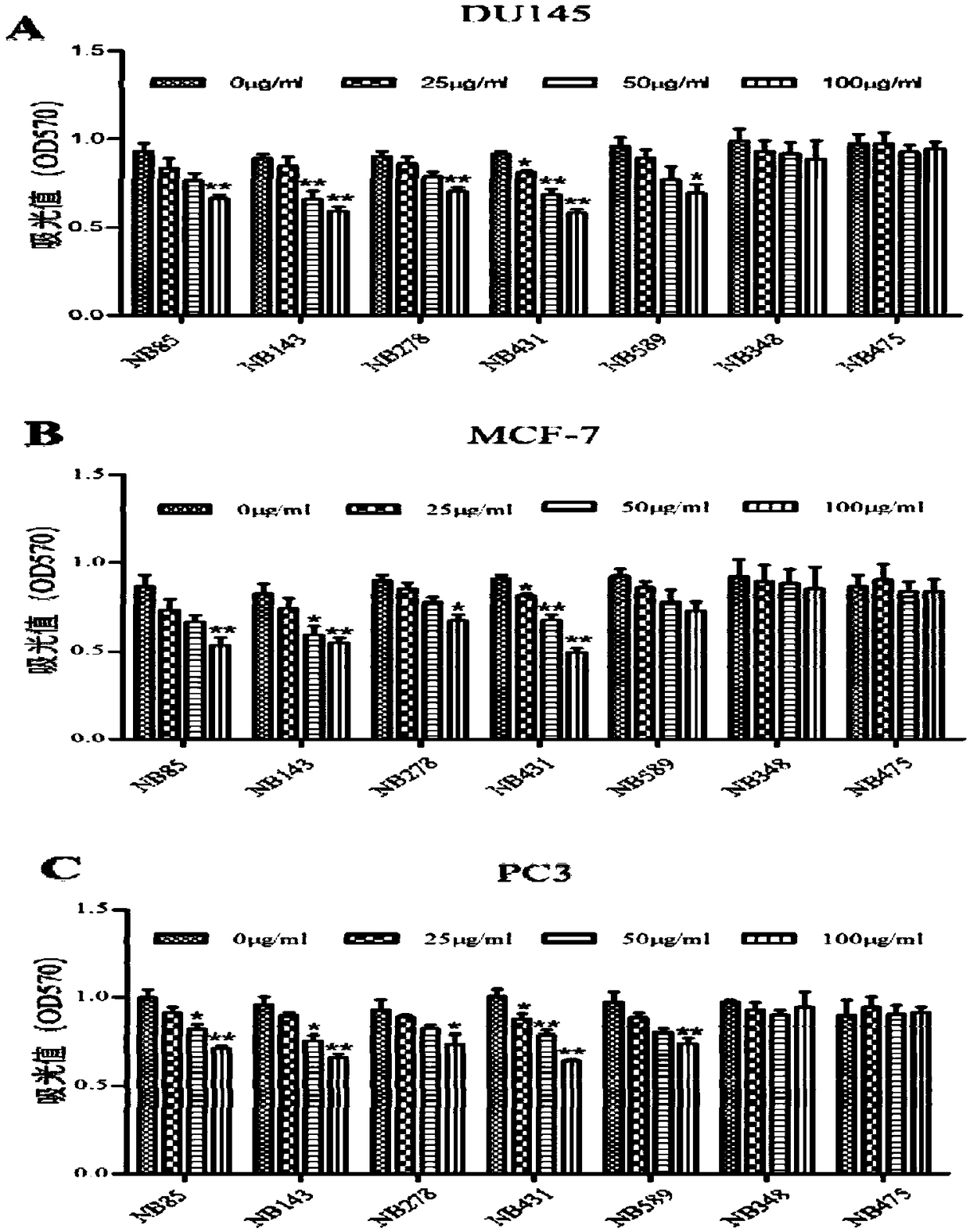
Binding protein of tumor stem cell marker molecule EpCAM and application of binding protein
A cancer stem cell and protein-binding technology, which is applied in the field of the binding protein of the tumor stem cell marker molecule EpCAM, can solve the problem of not being able to effectively kill tumor stem cells, and achieve the effects of easy transformation, reduced production costs, and accelerated application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
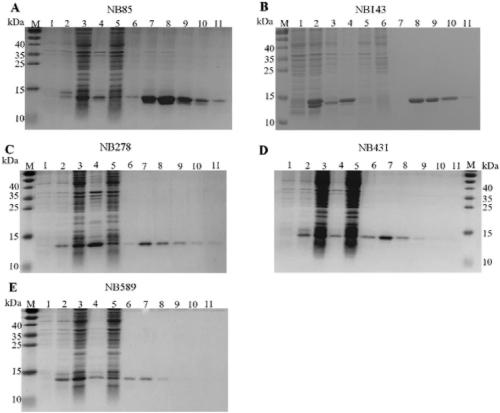
Examples
Embodiment 1
[0054] Example 1 preparation of helper phage
[0055] (1) Line the TG1 Escherichia coli clone strain in three zones on a petri dish with TYE solid medium, and place the marked petri dish upside down in a 37°C incubator for about 14 hours.
[0056] (2) Pick a single colony of TG1 on the petri dish and inoculate it into 5 mL of 2×TY liquid medium, and culture overnight at 37°C and 250 rpm.
[0057] (3) Transfer the bacterial solution obtained in step (2) to another 5mL 2×TY liquid medium at a ratio of 1:100 by volume, and culture it at 37°C and 250rpm until the bacterial solution OD 600 About 0.5.
[0058] (4) The KM13 helper phage was serially diluted with PBS (10 12 pfu / mL~10 4 pfu / mL).
[0059] (5) Take 200 μL of the bacterial solution cultured in step (3), add 10 μL of the diluted KM13 helper phage, and place the mixture in a 37° C. water bath for 30 minutes to obtain mixture A.
[0060] (6) Mix low-melting point agarose and 2×TY liquid medium, heat until completely mel...
Embodiment 2
[0066] Example 2 Preparation of phage binding protein library
[0067] (1) Thaw the TG1 bacteria containing binding protein particles (i.e. human binding protein phage library, SourceBioscience, London, UK) on ice, add 500mL 2×TY liquid medium (containing 100μg / mL ampicillin, 4% (w / v) Glucose), cultivated to OD at 37°C, 250rpm 600 = 0.5.
[0068] (2) add the 2 * 10 that embodiment 1 prepares 12 Helper phages were incubated in a water bath at 37°C for 30 minutes, 500 mL of the culture was divided into 50 mL tubes, centrifuged at 3200 g for 10 minutes, the supernatant was discarded, and the pellet was resuspended in 500 mL of 2×TY medium (containing 0.1% (w / v) glucose, 100 μg / mL ampicillin and 50 μg / mL kanamycin). 25°C and 250rpm shaking culture for 16-20h to obtain the phage binding protein library.
Embodiment 3
[0069] Example 3 Purification of the phage binding protein library
[0070] (1) The phage binding protein library prepared in Example 2 was divided into 50 mL tubes, 10 tubes in total.
[0071] (2) Centrifuge the centrifuge tube at 3200g for 20min, transfer the supernatant to a clean container, add 20% PEG-NaCl solution equivalent to 1 / 5 of the volume of the supernatant, mix well and distribute into 50mL centrifuge tubes Incubate on ice for 1 h.
[0072] (3) Centrifuge the centrifuge tube at 4°C, 3200g for 30min, discard the supernatant, resuspend the pellet with 5mL of PBS buffer, add 1mL of 20% PEG-NaCl solution to it, and incubate on ice for 10min.
[0073] (4) Centrifuge at 3200g at 4°C for 30min, discard the supernatant, resuspend the pellet with 1mL of PBS buffer, centrifuge at 3200g at 4°C for 5min, and filter the supernatant with a 0.45μM filter to sterilize.
[0074] (5) Roughly estimate the prepared phage titer by measuring the light absorption value at 260 nm: Dil...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com