NOVEL beta-1,6-ENDOGLUCANASE PRODUCING GENTIOBIOSE OR GLUCOSE FROM beta-GLUCAN
A technology for endoglucanase and gentiobiose, applied in the direction of glycosylase, enzyme, monosaccharide, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0041] obtain gly30b gene by cloning
[0042] will be derived from Proteus strain 2-40 T The putative β-1,6-endoglucanase (Sde_2994) of (ATCC43961) was cloned into Escherichia coli DH5α. A more specific description is as follows.
[0043] Proteus strains 2-40 at 30°C T (ATCC43961) was cultured for 12 hours in minimal medium containing 23g / L instant seawater salt, 50mM Tris-HCl, 2g / L glucose, 2g / L yeast extract, and 0.5g / L ammonium chloride.
[0044] Proteus strain 2-40 was obtained using a commercially available DNA isolation kit (Qiagen, Valencia, CA, USA). T (ATCC43961) genomic DNA.
[0045] The target gene gly30b (GenBank ID ABD82251.1) was amplified using Solg 2×Taq PCR Smart Mix 2 (SolGent, Daejeon, Korea). The primers used are as follows:
[0046] Forward primer: 5'-GCGGGATCCCACCACCACCACCACCACCAATACTGGTTAACCAGCGGTGATCTAAGT-3' (SEQ ID NO: 3); and
[0047] Reverse primer: 5'-GCGCTCGAGGTGGTGGTGGTGGTGGTGATCTATAACTAGCGTTACAACGCTCTGTGC-3' (SEQ ID NO: 4).
[0048] Thes...
example 2
[0050] Overexpression and purification of Gly30B protein
[0051] To overexpress the gene obtained according to Example 1, the gene was transformed into Escherichia coli BL21(DE3), which was a host for protein expression.
[0052] Use Luria-Bertani (Luria-Bertani; LB) medium (BD, Sparks, MD, USA) supplemented with 50 mg / L kanamycin (BD, Sparks, MD, USA)) at 37 The cells were incubated at °C until the absorbance at 600 nm reached 0.6. Protein expression was induced using 0.1 mM IPTG and the induction temperature was set at 16°C to express the recombinant protein in a water-soluble form.
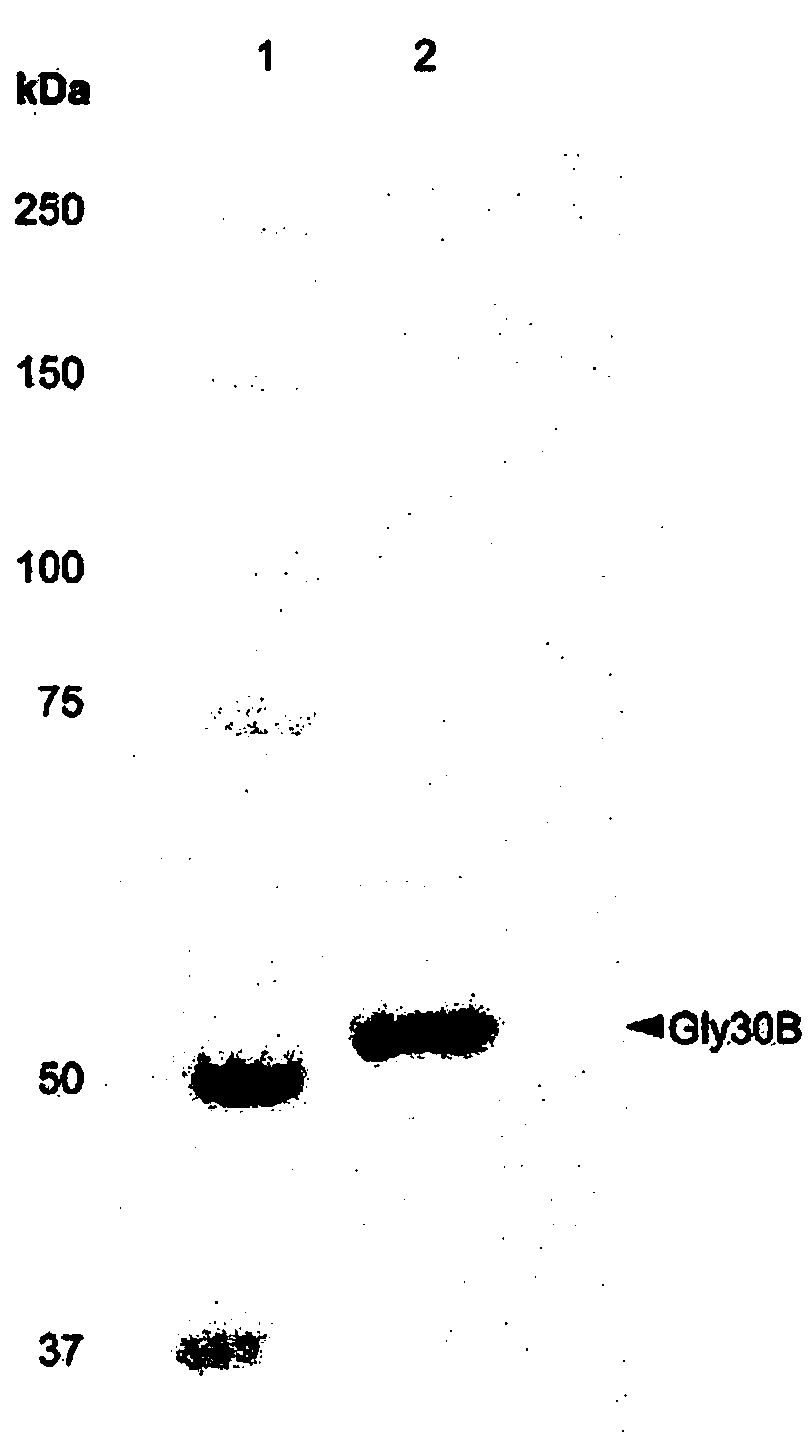
[0053] To isolate expressed Gly30B protein, cells were disrupted by sonication and centrifuged, and the supernatant was then purified using a HisTrap column (GE Healthcare, Piscataway, USA). Purified proteins were concentrated using Amicon Ultra centrifugal filters (Millipore, Billerica, MA, USA). The molecular weight of the expressed Gly30B protein was measured to be approximately 52 kDa ...
example 3
[0054] Verify the substrate specificity and cationic effect of Gly30B protein
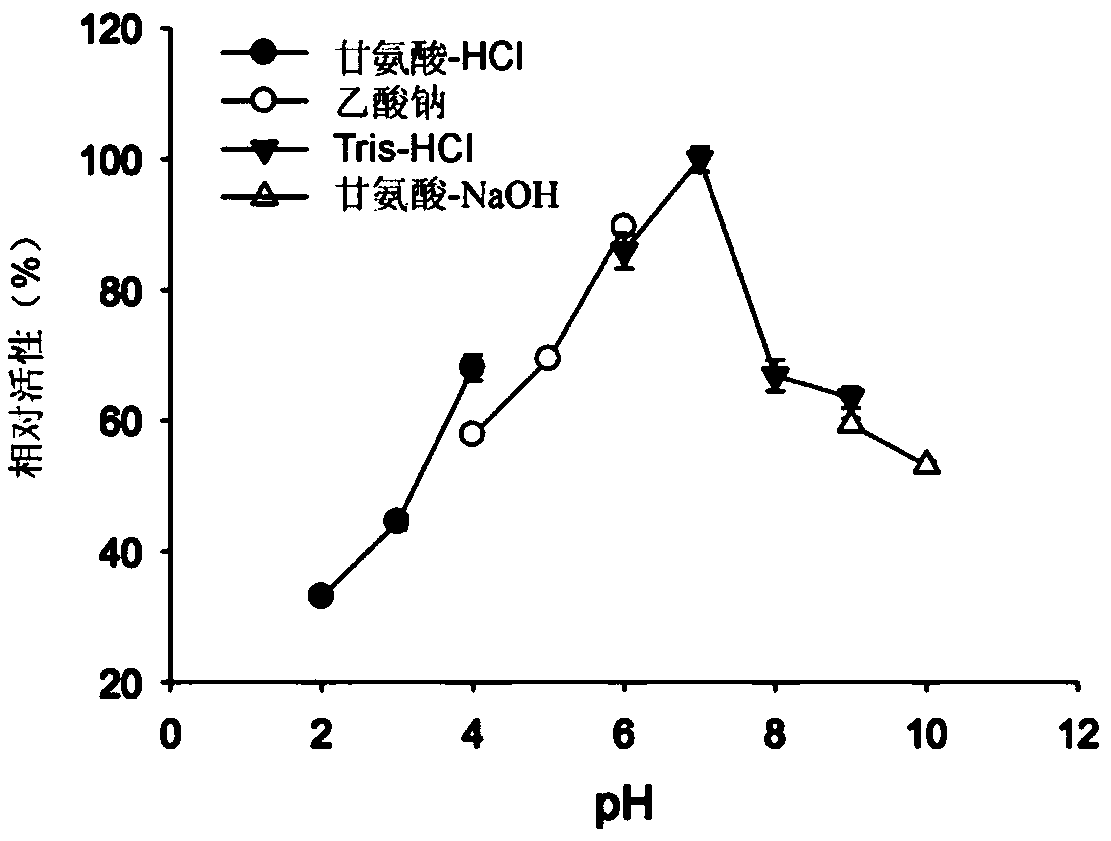
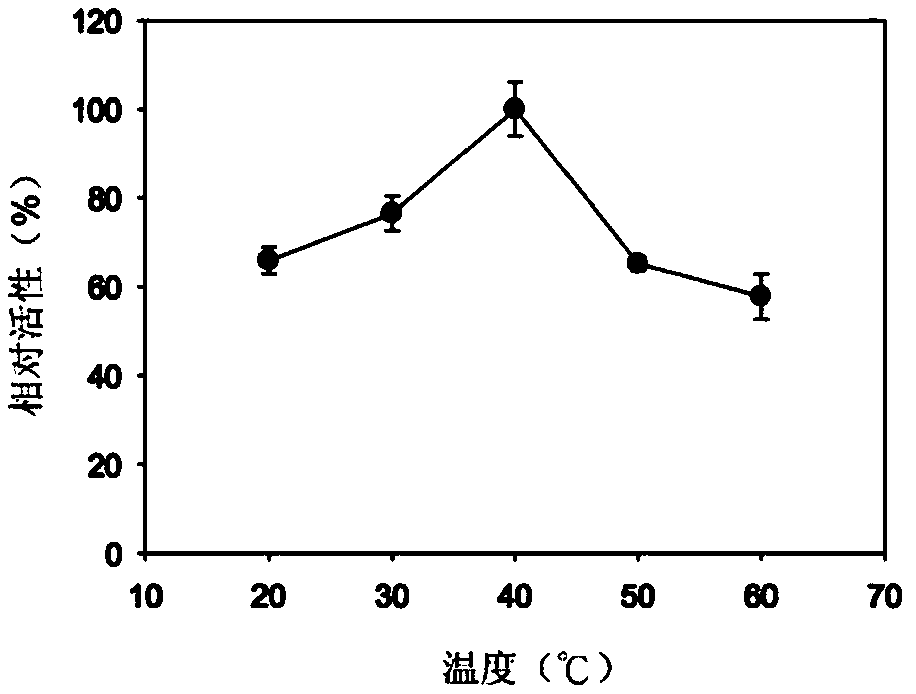
[0055] To confirm the enzymatic activity of the Gly30B protein, 1.89 nM Gly30B protein was allowed to dissolve in 100 μl of a matrix containing e.g. clamburin, laminarin (Wako, Osaka, Japan), etc. at 40°C at 2%. React in 20mM Tris-HCl (pH 6.0) for 30 minutes. In addition, in order to confirm the substrate specificity of Gly30B protein, 10.5 μM Gly30B protein was allowed to dissolve at 40°C in a mixture containing e.g. 100 μl of 20 mM Tris- React in HCl (pH 6.0) for 30 minutes. The reducing sugars produced were measured by the DNS method.
[0056] As shown in Table 1 below, the Gly30B protein showed the highest activity with respect to clamrose, and when laminarose was used as a substrate, the Gly30B protein exhibited about 22% relative activity. It was confirmed that the Gly30B protein does not hydrolyze curdlan, and this confirms that the Gly30B enzyme selectively cleaves β-1,6-glucan linkag...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com