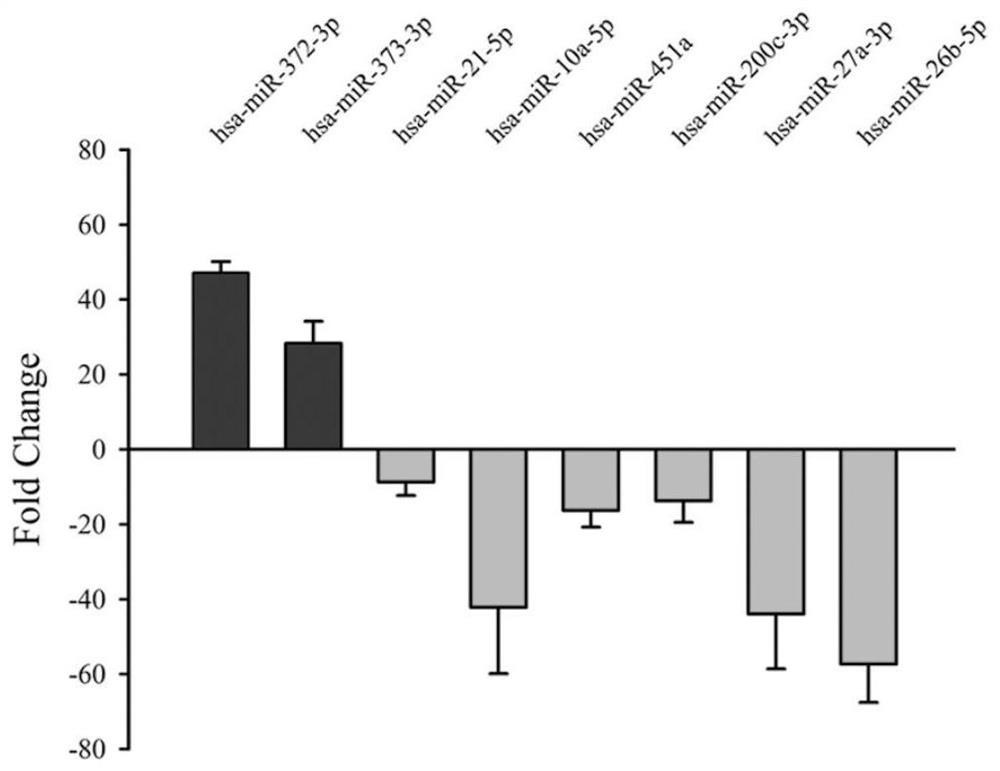
miRNA molecular markers for predicting the quality of early embryos in human assisted reproductive technology and their applications
A molecular marker, embryo technology, applied in the field of molecular biology, can solve the problem of no prediction, and achieve the effect of great social and economic benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] [Example 1] Sample collection and grouping
[0025] Embryo culture fluid samples were obtained from the Reproductive Medicine Center of Tongji Medical College of Huazhong University of Science and Technology, the Reproductive Center of Wuhan Union Medical College Hospital, and the Reproductive Center of Wuhan Tongji Hospital. The research was reviewed and approved by the Ethics Committee and informed consent was obtained from the patients. In the process of in vitro fertilization, oocytes are usually placed in 30-50ul droplets containing 5,000-10,000 sperm, and the sperm and eggs are co-cultured for 18-20 hours to allow in vitro fertilization to occur, observe the shape and number of pronuclei, and double-pronucleus The normal embryos were placed in the commercial embryo culture medium G1-plus (Vitrolife, Sweden), which is specially used for the growth and development of cleavage stage embryos. After continuous culture for 3 days, the culture medium used for IVF-ET embry...
Embodiment 2
[0026] [Example 2] Extraction of total RNA and miRNA library construction
[0027]Total RNA was extracted from the embryo culture fluid specimens of group A and group B respectively, and the total RNA extracted was quantified and inspected by Nanodrop instrument (Thermo Company, USA). Add 3' and 5' adapters to the RNA respectively, amplify by RT-PCR, and recover the target band of 145-160 bp by electrophoresis to obtain the miRNA library.
Embodiment 3
[0028] [Example 3] RNA-Seq high-throughput sequencing to screen differential miRNAs
[0029] Qubit (Invitrogen, USA) quantified the miRNA library, used the Start cBot instrument for cluster generation, and then sequenced with Hiseq2500 (Illumnia, USA) to obtain raw data, and calculated miRNA expression. Use DESeq software to analyze the differential expression of the two groups of samples, calculate the expression level, the mean value within the group and the differential expression multiple between the groups, and further calculate the log2 (difference multiple) value, when the P value between the two groups is ≤0.05 and the log2 (difference multiple) )≥1, it is considered that the expression of the miRNA is different between the two groups. Through the above differential expression analysis of the sequencing data, combined with the GO clustering analysis of the specific biological functions of miRNA target genes, 18 differentially expressed miRNAs related to biological func...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com