SNP molecular markerand method for identifying mycobacteria, primer composition, kit and application
A technique of primer composition and mycobacteria, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of long time, easy false positives in picture results, low resolution, etc., to improve accuracy degree of effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
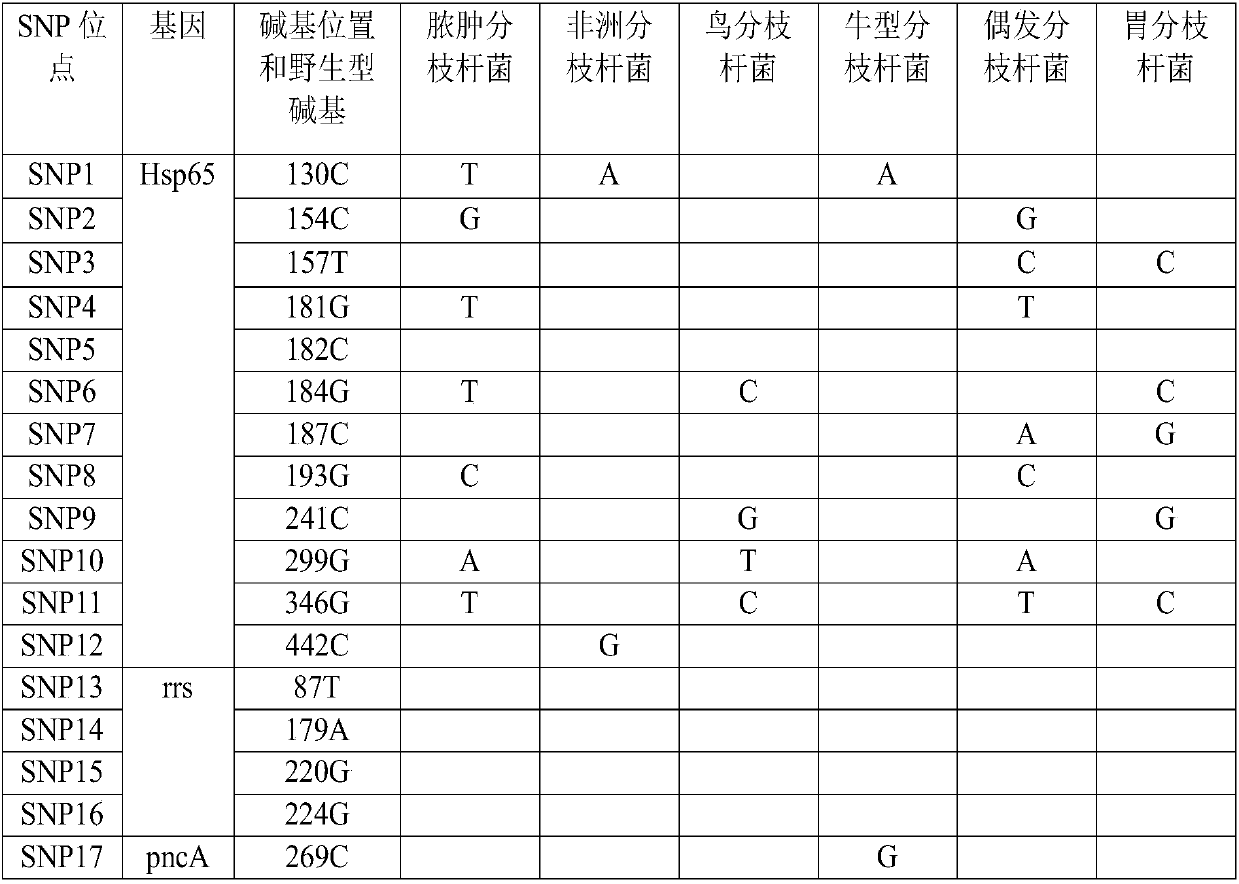
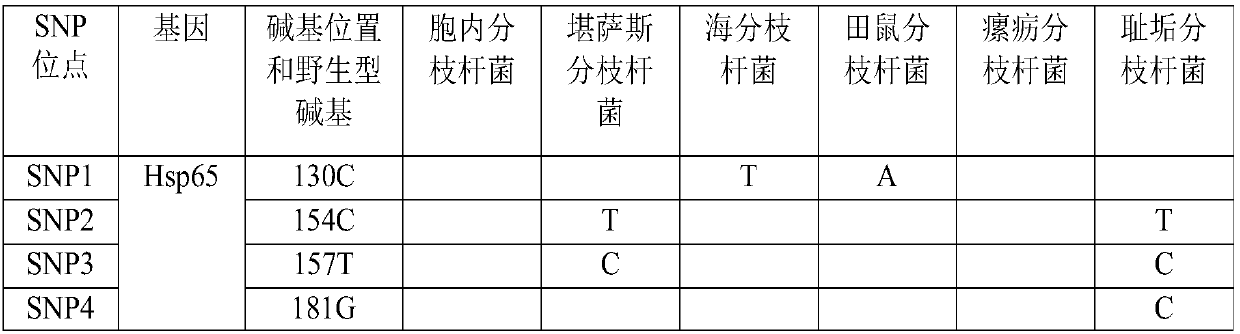
[0087] This embodiment provides a primer composition for identifying mycobacteria, including a primer pair Myco1, a primer pair Myco2 and a primer pair Myco3, specifically as shown in Table 4.
[0088] Table 4 primer pair Myco1, primer pair Myco2 and primer pair Myco3
[0089]
Embodiment 2
[0091] Using the primer composition provided in Example 1 to identify 18 kinds of mycobacteria, including Mycobacterium tuberculosis M.tuberculosis (ATCC 27294) belonging to the Mycobacterium tuberculosis complex, Mycobacterium bovis M.Bovis (ATCC19210) and Tian Mycobacterium murine M. Microti (ATCC 19422); also includes Mycobacterium avium M. avium (ATCC 25291 ), which belongs to nontuberculous mycobacteria (MOTT) outside the Mycobacterium tuberculosis complex. All strains were strains preserved in the Beijing Tuberculosis Clinical Data and Sample Resource Bank of Beijing Chest Hospital Affiliated to Capital Medical University.
[0092] With the genomic DNA of test bacterial strain as template, adopt the primer pair that embodiment 1 designs to carry out PCR amplification.
[0093] PCR reaction: use the three sets of primer pairs in Table 4, and adopt a conventional PCR reaction system. The reaction system is as follows: the total volume is 20 μl, including 2×Premix Taq (Code...
Embodiment 3
[0098] The present embodiment provides a method for identifying mycobacteria, comprising the steps of:
[0099] 1. For the collection of clinical sputum samples, use the QIAamp DNA Mini Kit (CAT.No.51304) kit to extract DNA through pretreatment;
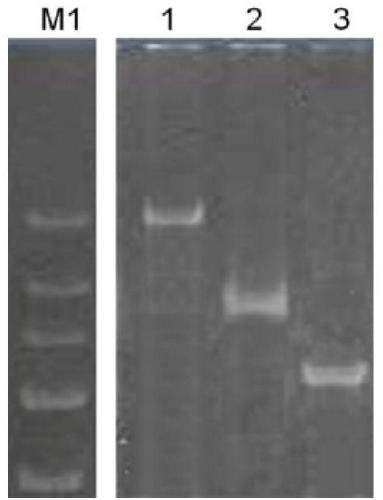
[0100] 2.PCR reaction: use the primer pair of three groups in table 4, adopt conventional PCR reaction system, the reaction system is as follows: the total volume is 20 μ l, including 2×Premix Taq (Code No.: R004A, Takara company, Premix is made by DNAPolymerase , Buffer, dNTP Mixture) 10 μl, 10 μM upstream and downstream primers 1 μl (final concentration 0.5 μM), DNA template 1 μl, double distilled water 7 μl to 20 μl system.
[0101] Reaction conditions: 94°C for 5min, 30 cycles: 94°C for 45sec, 60°C for 45sec, 72°C for 50sec, and finally extension at 72°C for 7min. The same annealing temperature is used for the primers, which reduces the operation steps and saves the time for strain identification.
[0102] PCR results such as...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com