Construction of EP153R and EP402R gene co-expression recombinant adenovirus vector and adenovirus packaging method
A technology of EP402R and EP153R, applied in the direction of virus/phage, microorganism-based method, vector, etc., can solve the problem of no EP153R recombinant adenovirus vector, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] i. Optimized synthesis of the gene in Example 1
[0038] EP153R gene synthesis: query the EP153R gene (NC_001659.2), EP402R gene (NC_001659.2) and CTLA4 gene (NC_010457.5) from pig cell lines included in the NCBI website, and obtain optimized (referring to nucleoside optimized acid sequence), the sequence is as SEQ ID No.1-3.
[0039] CTLA4 sequence Seq ID No.1
[0040] GCTAGAGATCTGGTACCGCCACCATGCACGTGGCCCAACCTGCAGTAGTGCTGGCCAACAGCCGGGGTGTTGCCAGCTTTGTGTGTGAGTATGGGTCTGCAGGCAAAGCTGCCGAGGTCCGGGTGACAGTGCTGCGGCGGGCCGGCAGCCAGATGACTGAAGTCTGTGCCGCGACATATACTGTGGAGGATGAGTTGACCTTCCTTGATGACTCTACATGCACTGGCACCTCCACCGAAAACAAAGTGAACCTCACCATCCAAGGGCTGAGAGCCGTGGACACTGGGCTCTACATCTGCAAGGTGGAGCTCCTGTACCCACCACCCTACTATGTGGGTATGGGCAACGGGACCCAGATTTATGTCATTGATCCAGAACCATGCCCAGATTCTGATAGTACTGATTACAAAGACGATGACGATAAG
[0041] EP153R sequence Seq ID No.2
[0042] AAAGACGATGACGATAAGACCGGTTTCAGCAACAAGAAGTACATCGGCCTGATCAACAAGAAGGAGGGCCTGAAGAAGAAGATCGATGACTACAGCATCCTGATCATCGGCATCCTGATCGGCACCAACATCCTGAG...
Embodiment 3
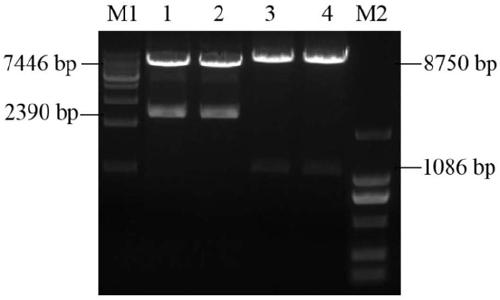
[0063] iii. Example 3 Restriction digestion and homologous recombination of pShuttle-CMV vector
[0064]1) Digest the pShuttle-CMV vector, and the restriction system is shown in Table 3.
[0065] Table 3 enzyme digestion system
[0066] Reaction solution composition
volume
Plasmid (350ng / μL)
20 μL
10×Buffer
5μL
KpnI
2μL
Hind III
2μL
ddH2O
31μL
total
50μL
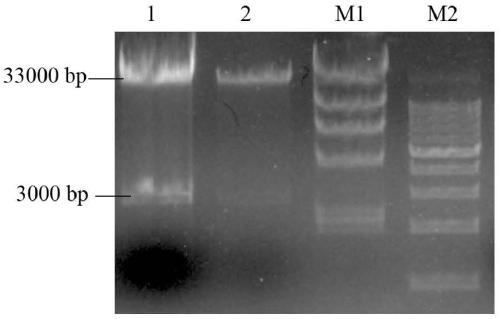
[0067] After adding the sample and mixing it, place it at 37°C for enzyme digestion for 6 hours. After the reaction, use 1% agarose gel electrophoresis to detect the size of the digested band, and use a gel recovery kit to recover the linearized plasmid.
[0068] 2) Vector and fragment homologous recombination:
[0069] Table 4 Reaction system for homologous recombination of vectors and fragments
[0070] Reaction solution composition
volume
pShuttle-CMV (90ng / μL)
2μL
CTLA4-EP153 (50ng / μL)
1.5μL
EP...
Embodiment 4
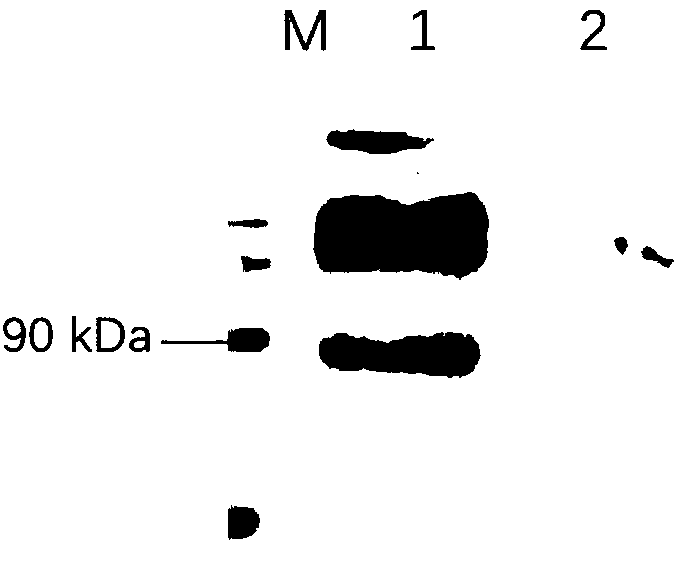
[0076] iv. Example 4 Transient expression of 293T cells and its verification
[0077] 1) 293T cell transfection
[0078] Cell preparation: 1 x 10 6 293T cells were evenly spread in each well of a 6-well plate, and used for cell transfection after the cells adhered to the wall the next day;
[0079] Plasmid preparation: Mix 2.5 μg pShuttle-CMV-CTLA4-EP153R-EP402R plasmid with 7.5 μL Lip3000 liposomes and 500 μL serum-free DMEM medium, let stand for 4 minutes, mix the plasmid mixture with Lip3000 mixture, let stand 10min;
[0080] The mixture was added dropwise to the culture medium, shaken gently, and cultured at 37°C.
[0081] Cells were collected after 24 h.
[0082] 2) WB detection of protein expression
[0083] Protein extraction: wash twice with PBS, add 250μl cell lysate (containing protease inhibitors and protein phosphatase inhibitors), digest and blow off the cells, shake and mix, ice bath for 25min, centrifuge at 12000rpm for 15min, absorb 200μl supernatant and a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com