Method for positioning rice complex trait gene
A technology of gene positioning and rice, applied in the direction of genomics, biochemical equipment and methods, microbial measurement/inspection, etc., to achieve the effect of low cost, high efficiency and short time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
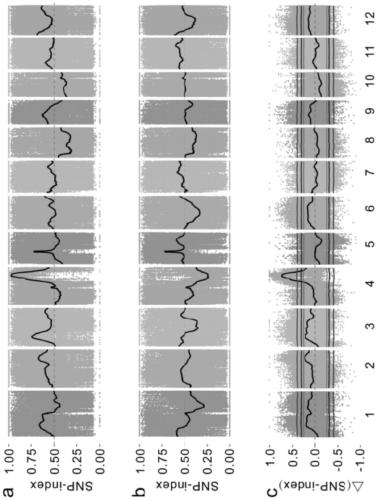
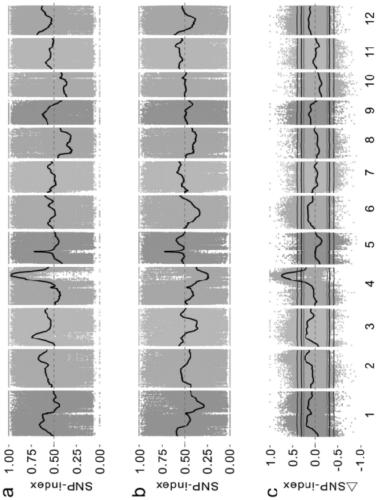
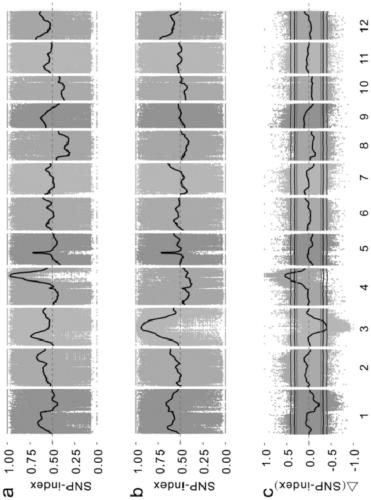
[0036]A method for gene mapping of rice complex traits, comprising the following steps:
[0037] (1) Population construction: Donglanmo with black seed coat and anthocyanin content as high as 1642.99 μg / g and Huang Huazhan with white seed coat and as low as 3.84 μg / g anthocyanin were selected as parents. F 2 group;
[0038] (2) Phenotype identification and grouping: the parents Huang Huazhan, Donglan Momi and F of the detection step (1) 2 The seed coat color of each individual plant in the population was determined by high performance liquid chromatography, and the anthocyanin content of brown rice was determined. According to the seed coat color and anthocyanin content of each individual plant, the F 2 The population is divided into 4 subgroups: completely black, partially black, brown, white;
[0039] The above-mentioned brown rice anthocyanins include 6 kinds of anthocyanins: geranium pigment, morning glory pigment, delphinium pigment, paeoniflorin pigment, cyanidin pigm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com