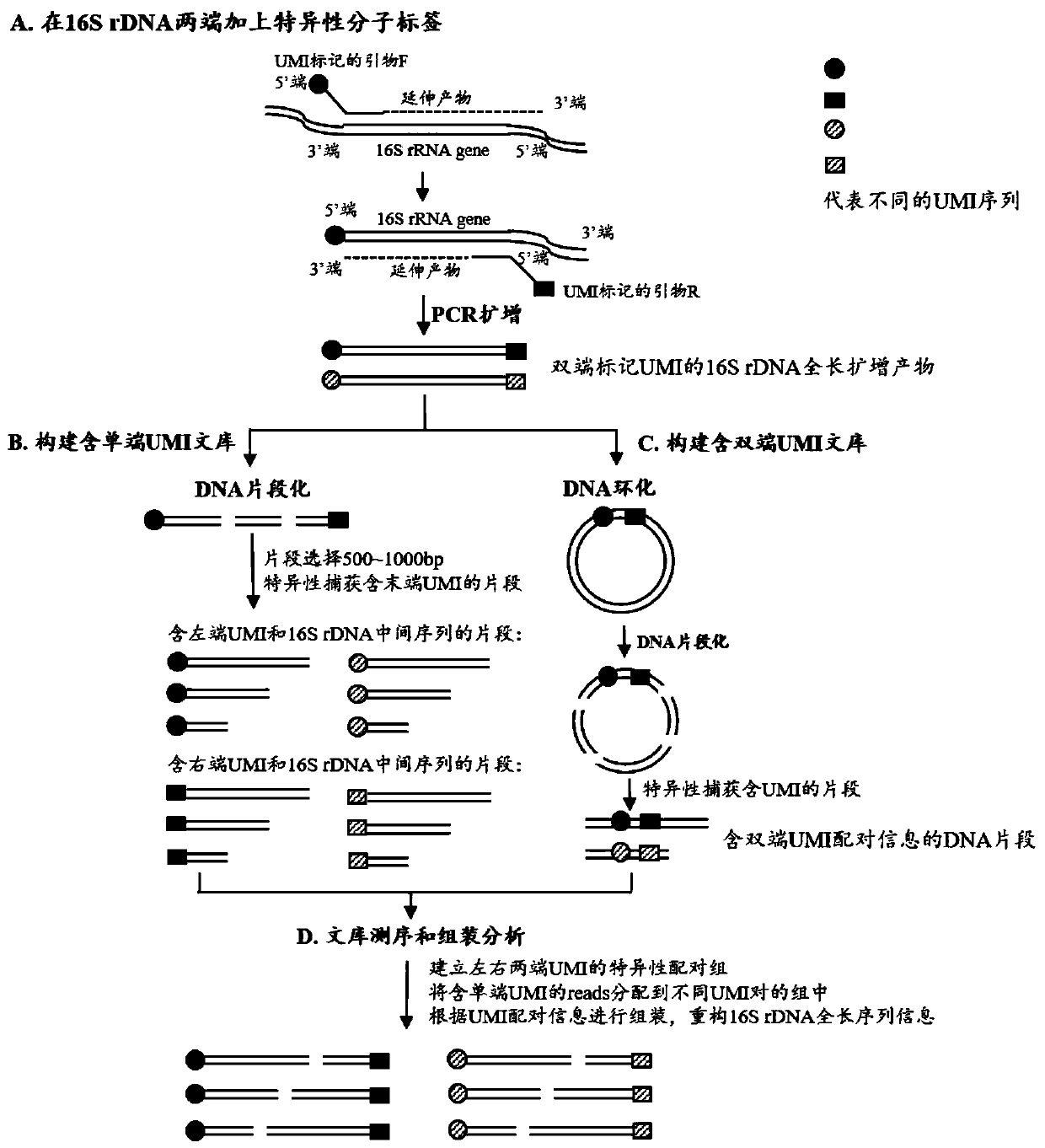
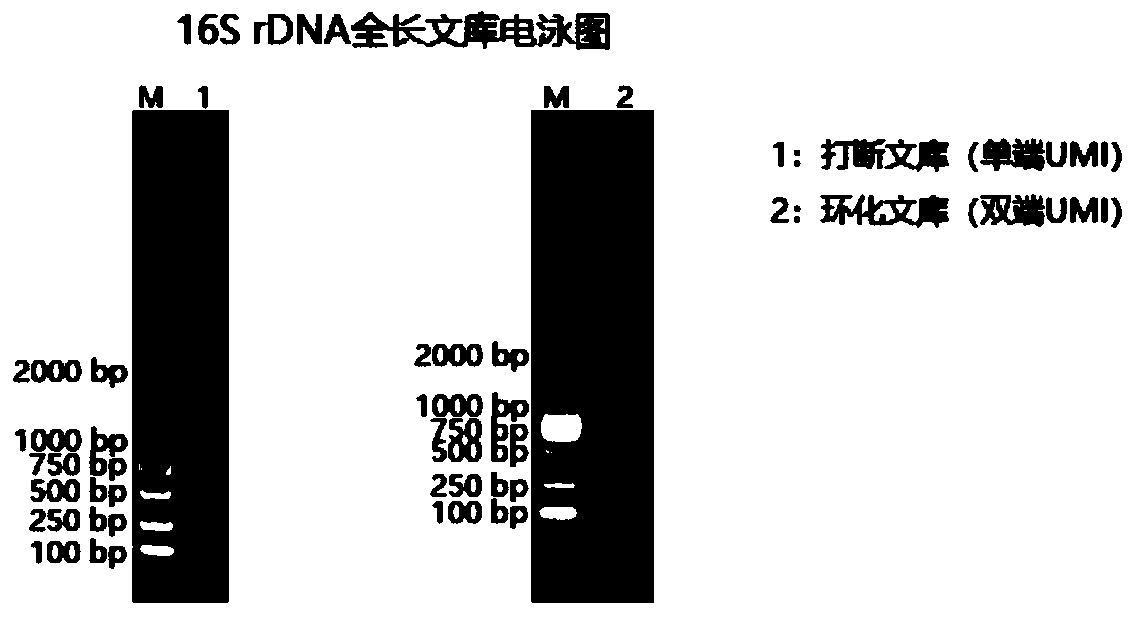
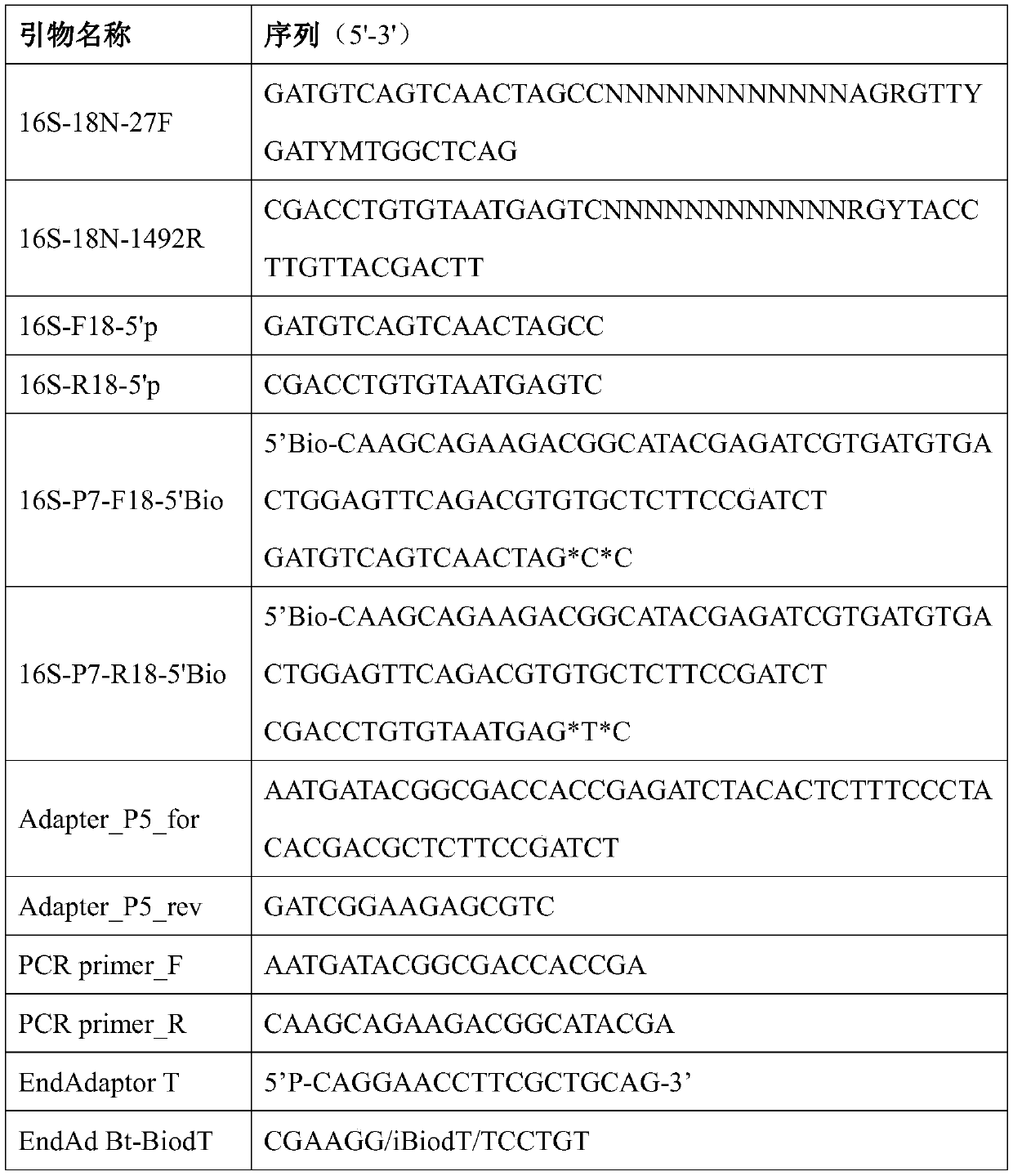
Method for constructing bacterium 16S rDNA overall-length high-throughput sequencing library
A high-throughput, library-based technology that can be used in chemical libraries, biochemical equipment and methods, and microbial assays/examinations. It can solve the problems of low throughput and high sequencing costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0057] The following specific examples are further descriptions of the methods and technical solutions provided by the present invention, but should not be construed as limiting the present invention.
[0058] 1. Primer Design
[0059] The primer sequence information used in this embodiment is as follows:
[0060]
[0061] Note: * stands for thio modification; Bio stands for biotin modification; N stands for random base, each random base N is selected from any one of A, T, C, and G.
[0062] 2. Extraction of Bacterial Genomic DNA from Samples
[0063] Take an appropriate amount of human feces samples, and use the Fecal Genomic DNA Extraction Kit of Tiangen Biochemical Technology Co., Ltd. to extract the genomic DNA of intestinal bacteria. The intestinal bacteria genome DNA was taken, and mixed with 15% Stenotrophomonas bacterial DNA and 5% Chryseobacterium bacterial DNA according to the mass ratio. The mixed DNA was evenly divided into two parts, one of which was used to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com