Ribosome DNA methylation marker for detection of cancer in peripheral blood and application thereof
A marker, ribosome technology, applied in recombinant DNA technology, DNA/RNA fragment, microbial determination/inspection, etc., can solve the problems of poor marker specificity, loss of radical resection, etc., to achieve high specificity and sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Genome-wide methylation sequencing data screening of differential CpG sites on ribosomal DNA
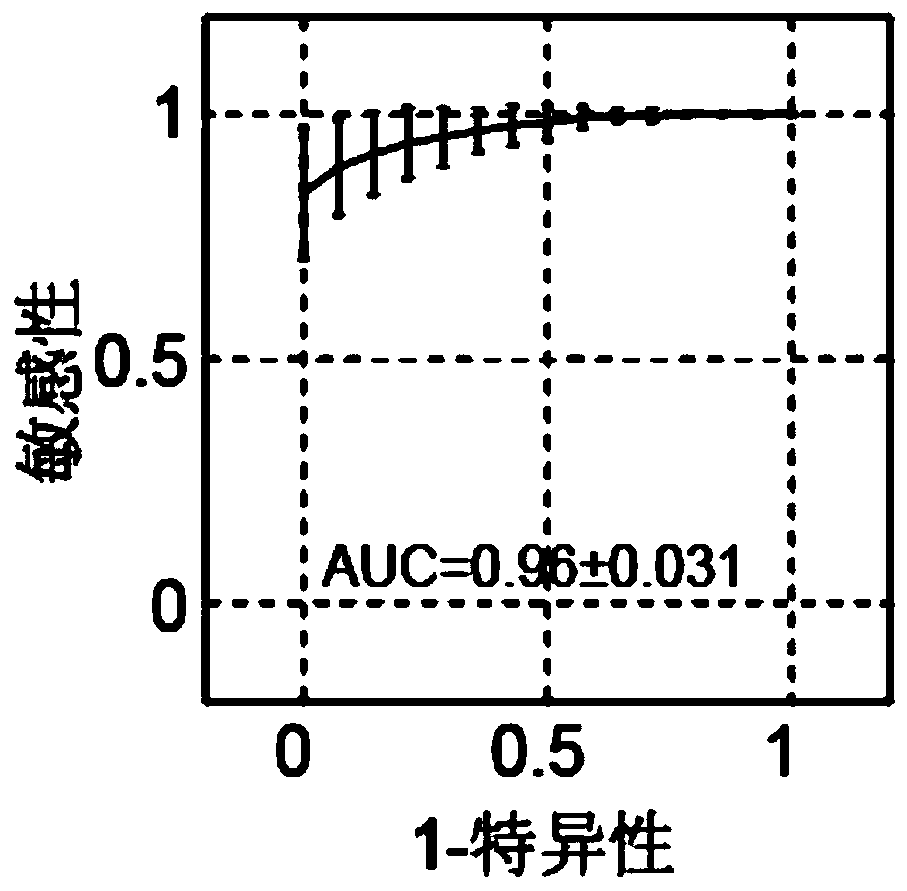
[0057] We used peripheral blood bisulfite sequencing data published in PNAS titled "Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasmaDNA bisulfite sequencing" in 2013, and the data were deposited in the European Genome-Table Phenome Archives (European Genome-Phenome Archive), accession number is EGAS00001000566. Here we use the peripheral blood DNA methylation data of healthy people (32), HBV-infected non-cancer patients (8), early liver cancer patients (stage I, stage II, 26), and 15 pairs of liver cancer tissues and white blood cells Layer DNA methylation data.
[0058] Download the reference sequence U13369.1 of the human ribosomal DNA repeat fragment unit in GENBANK, with a total length of 42999 bases and 3288 CpG sites. Use the bs-seeker2 software to match the sequencing data back to U13369.1 on the re...
Embodiment 2
[0065] Example 2 Using the model and screening sites obtained in Example 1 to predict the effect of lung cancer, breast cancer, and nasopharyngeal cancer
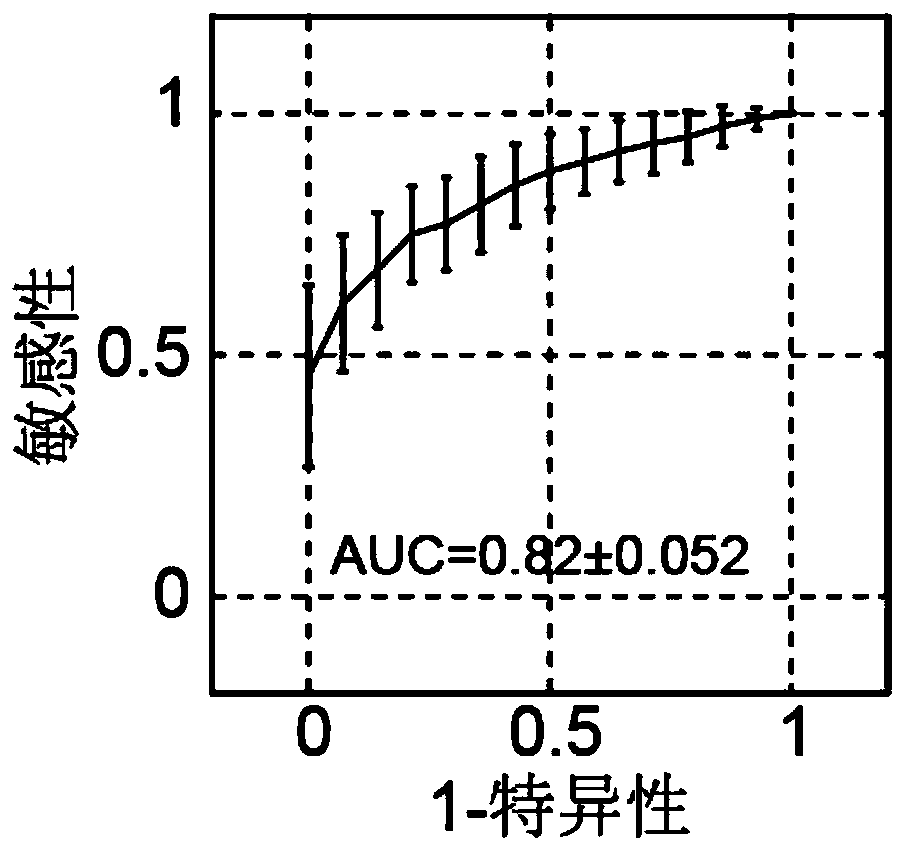
[0066] The data used the reference data given in Example 1, including 5 breast cancers, 4 lung cancers, and 9 nasopharyngeal carcinomas. The number of cancer patients here is limited, and these types of cancer do not distinguish between cancer types. The models and loci obtained from liver cancer training in Example 1 were used to predict the distinction between these three types of cancer and reserve healthy people. The purpose of this example is to verify that the sites and models screened in Example 1 can also be used for the detection of other cancer types. figure 2 Gives a plot of the predicted ROC curve. From figure 2 It can be seen that the average AUC reaches 0.82, which has achieved a good prediction effect. It illustrates that the sites screened in Example 1 can be used for the prediction of breast cancer, l...
Embodiment 3
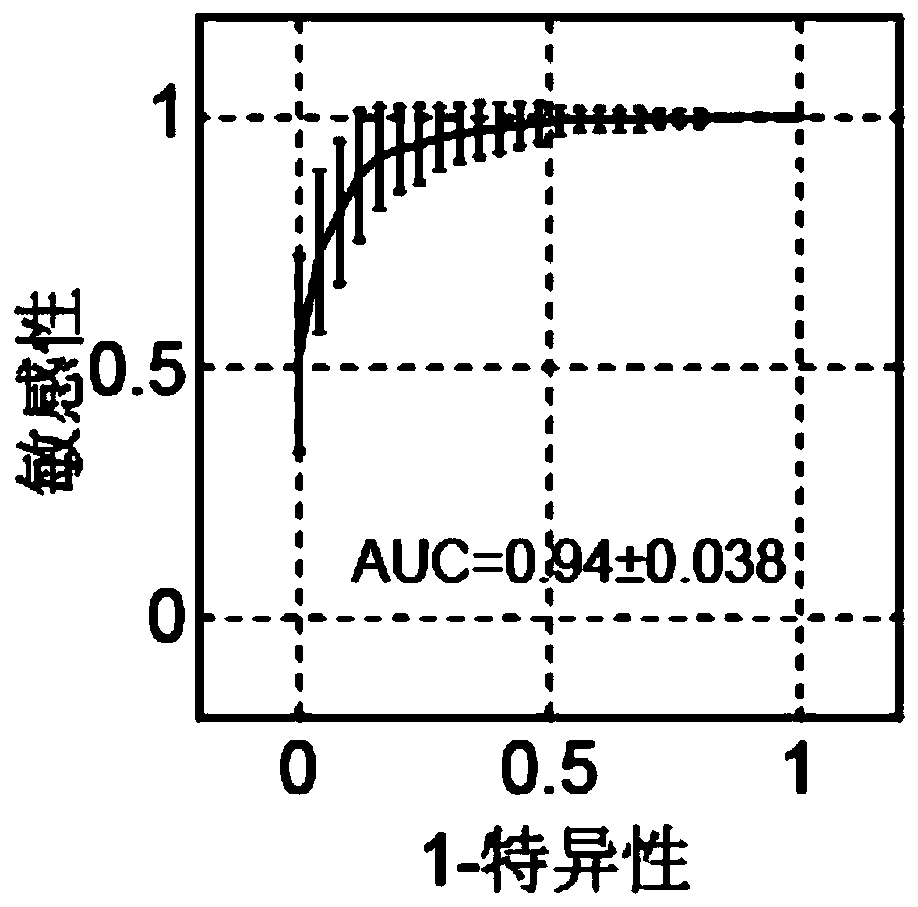
[0067] Example 3 Using Peripheral Blood RRBS Data to Predict Lung Cancer and Colorectal Cancer
[0068] We used the degenerate representative bisulfite sequencing (RRBS) of peripheral blood published in the PNAS article titled "Identification of methylation haplotypeblocks aids in deconvolution of heterogeneous tissue samples and tumorissue-of-origin mapping from plasma DNA" in 2017 Data, data deposited in Gene Expression Omnibus (GEO), accession number is GSE79279. Here, the peripheral blood DNA methylation data of healthy people (75), lung cancer patients (29), and peripheral blood DNA methylation data of 30 colorectal cancer patients were used.
[0069] Download the reference sequence U13369.1 of the human ribosomal DNA repeat fragment unit in GENBANK, with a total length of 42999 bases and 3288 CpG sites. Use the bs-seeker2 software to match the sequencing data back to U13369.1 on the reference sequence, and no longer remove the sequencing repeats, because the sequencing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com