Method for detecting copy number variation by means of single sample based on second-generation sequencing technology, and computer system
A second-generation sequencing technology and copy number variation technology, applied in the field of methods and computer systems, can solve problems such as high cost of experiments and analysis, cumbersome and complicated overall processes, and achieve effective correction of false positives and false negatives, simplifying experiments and analysis steps, high consistency effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
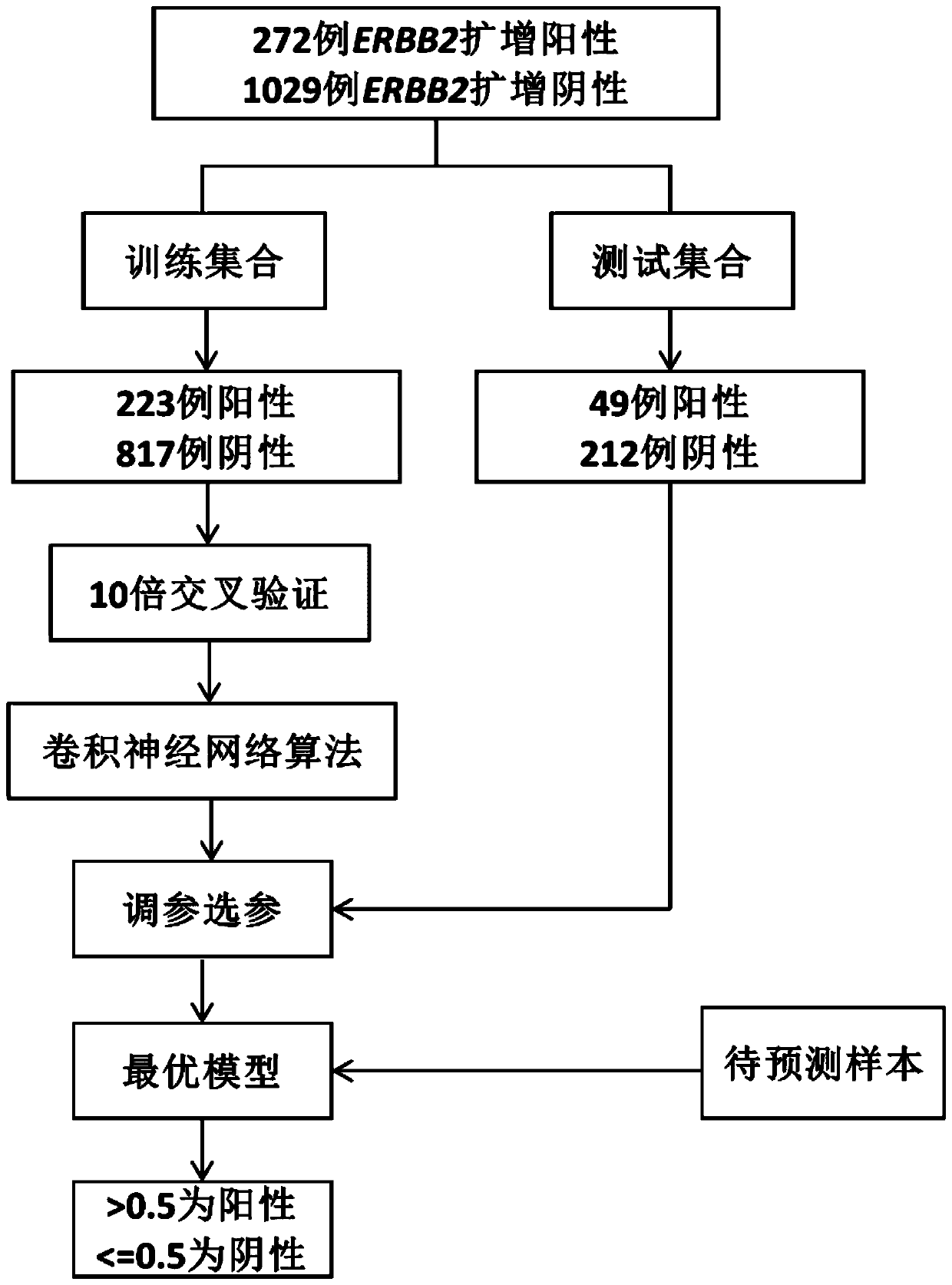
[0027] The whole exome data of 5 known ERBB2 amplification-positive cases and 5 known ERBB2-amplified negative cases were selected to test the analysis method of the present invention. Specifically take 1 example of ERBB2 positive sample embodiment Sample1 as an example (such as flow process figure 1 ), all the other embodiments repeat steps 1-11, as follows:
[0028] 1. Collect the whole exome sequencing data of 272 cases of positive ERBB2 gene amplification, and collect the whole exome sequencing data of 1029 cases of negative ERBB2 gene amplification, and divide the data into two parts: training set and test set; The training set includes 223 positive samples and 817 negative samples, and the test set includes 49 positive samples and 212 negative samples;
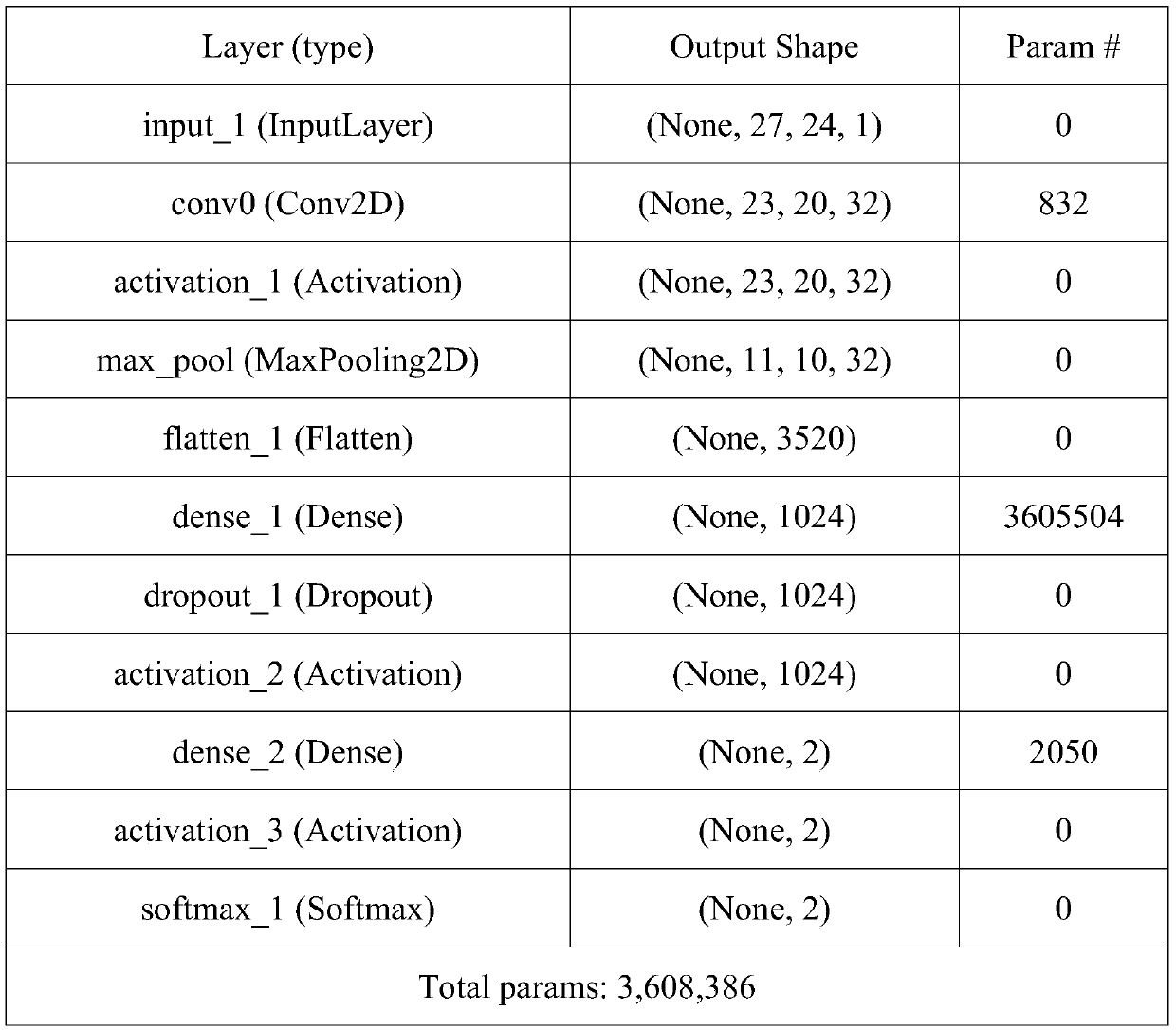
[0029] 2. The gene ERBB2 contains 27 full exons. The 27 amplified or deleted regions Lj (0
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com