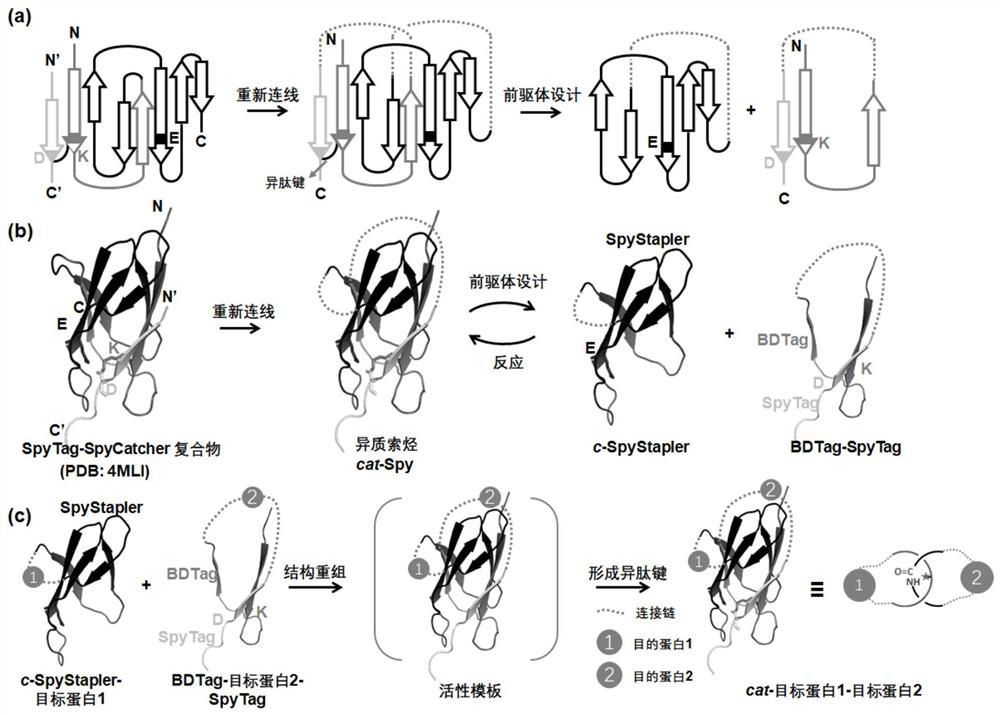
A protein coupling method based on catenation
A protein and coupling technology, applied in the field of preparation of biomacromolecular proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1: Expression and purification of reaction precursor protein
[0058] During the construction process, the reaction precursors BD-A, BD-ELP2-A and BD-DHFR-A were constructed in the pQE-80L vector, and c-S, c-S-E1, c-S-GFPrm were constructed in the pET-15b vector. The vector was transformed into Escherichia coli BL21(DE3) competent cells and cultured overnight. Then select the single clone grown on the plate, inoculate into 5 mL LB medium containing 100 μg / mL ampicillin sodium, and culture at 37° C. for 8-12 hours. Then transfer the obtained seed liquid into 1L of fresh LB medium at a ratio of 1:100 or 1:200, and culture it with shaking at 37°C until OD 600 Between 0.6-0.8, add isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and transfer to 16°C for expression for 20 hours. Bacteria were collected by centrifugation. Then the cells were dispersed in 30-35mL lysis buffer A (50mM disodium hydrogen phosphate, 300mM sodium chloride, 10m...
Embodiment 2
[0059] Embodiment 2: the characterization of reaction precursor
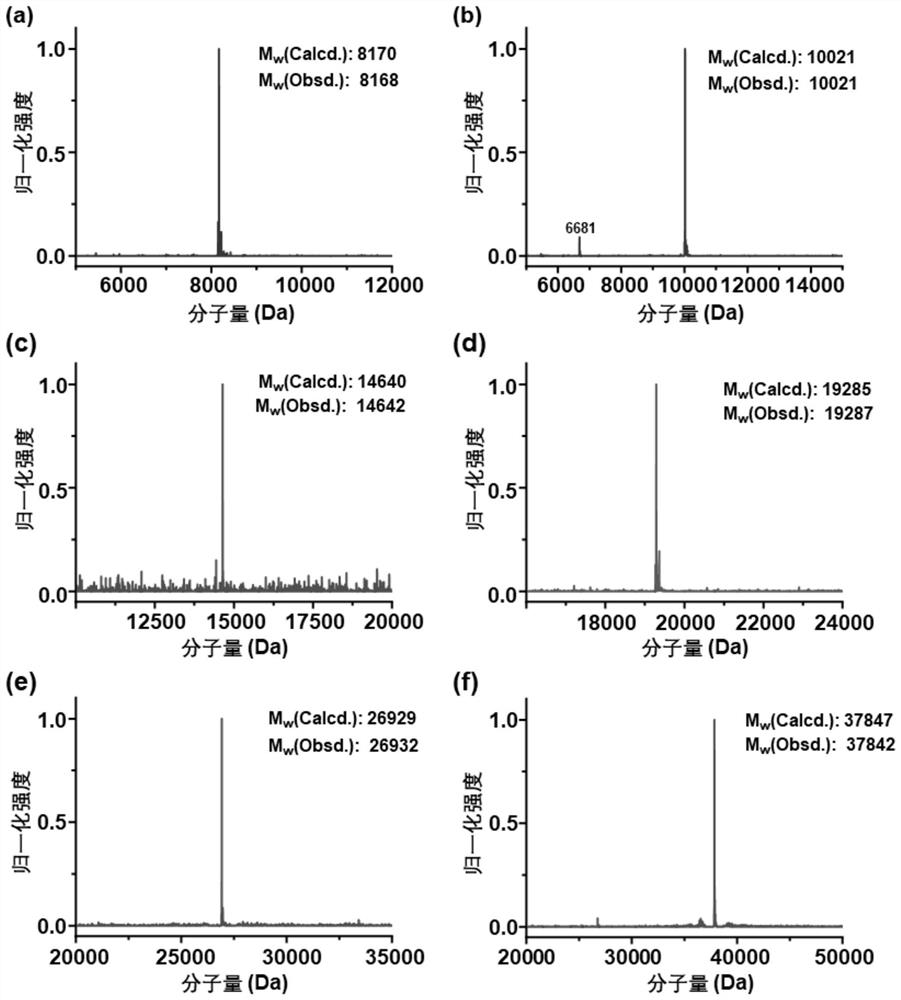
[0060] The purified reaction precursor was initially characterized by high performance liquid chromatography-electrospray mass spectrometry. figure 2 Precursors BD-A (a), c-S (b), BD-E2-A (c), c-S-E1 (d), BD-DHFR-A (e), and The mass spectrometry data of c-S-GFPrm(f), the experimental data and the theoretical calculation data are basically consistent.
Embodiment 3
[0061] Example 3: Preparation and Characterization of Protein Heterogeneous Catanenes
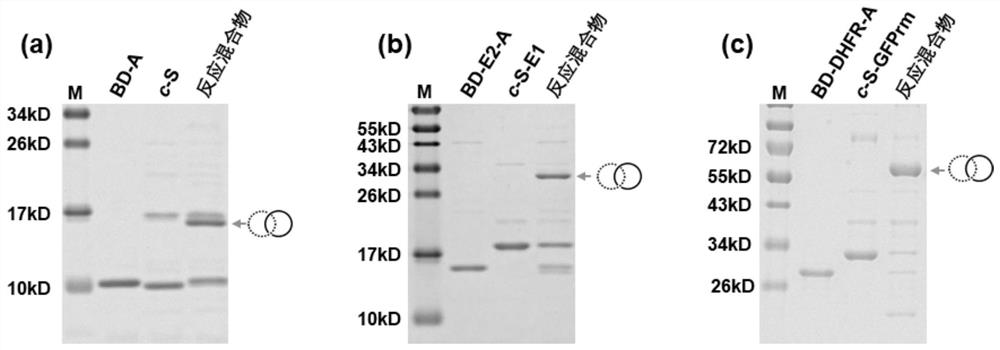
[0062] The purified reaction precursors BD-A / BD-E2-A / BD-DHFR-A were mixed with c-S / c-S-E1 / c-S-GFPrm at a concentration ratio of 1:1 at a reaction concentration of 50 μM. 10mM dithiothreitol (DTT) (5mM DTT was added to the BD-DHFR-A and c-S-GFPrm reaction system) was used to reduce and separate the intermolecular disulfide bonds. The mixed system was reacted at 4°C for 20h. After the reaction, 10 μL of the reaction precursor was mixed with the reaction mixture and 5× protein loading buffer, then heated at 98°C for 10 min, and the results were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. image 3 The reaction conditions of the three pairs of reaction precursors are shown, and new bands with molecular weights corresponding to the sum of the two reaction precursors appear in the reaction mixture, that is to say, a catenane structure is formed. Afterwards, the reactio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com