A method for determining the mechanism of action of antibiotics using a bioluminescent reporter system
A reporting system, bioluminescence technology, applied in chemiluminescence/bioluminescence, introduction of foreign genetic material using vectors, material analysis by optical means, etc. There are no problems such as the mechanism of action of antibiotics, time-consuming and labor-intensive, etc., to achieve the effect of improving the speed of identification, huge social and economic benefits, and improving the speed of discovery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Experimental strains and plasmids:
[0036]Escherichia coli DH5α: used for competent preparation, obtained from China General Microorganism Culture Collection Center (CGMCC1.12873).
[0037] Escherichia coli DH10B: used for competent preparation, obtained from ThermoFisher Scientific (Cat. No. 18297010)
[0038] Staphylococcus aureus RN4220: used for competent preparation and luminescence detection, acquired from BEI Resources.
[0039] pMD19-T: Purchased from Dalian Bao Biological Company (TaKaRa)
[0040] pAmilux: from the University of British Columbia, Canada (Document: Mesak LR, Yim G, Davies J, Improved lux reporters for use in Staphylococcus aureus, Plasmid, 2009, 61(3): 182-187.)
[0041] The medium and its preparation required for the experiment:
[0042] LB medium: For each liter of medium, tryptone (10g), sodium chloride (10g), and yeast extract (5g) should be added to 950ml of deionized water, and stirred with a magnetic stirrer until dissolved. Add 5 mo...
Embodiment 2
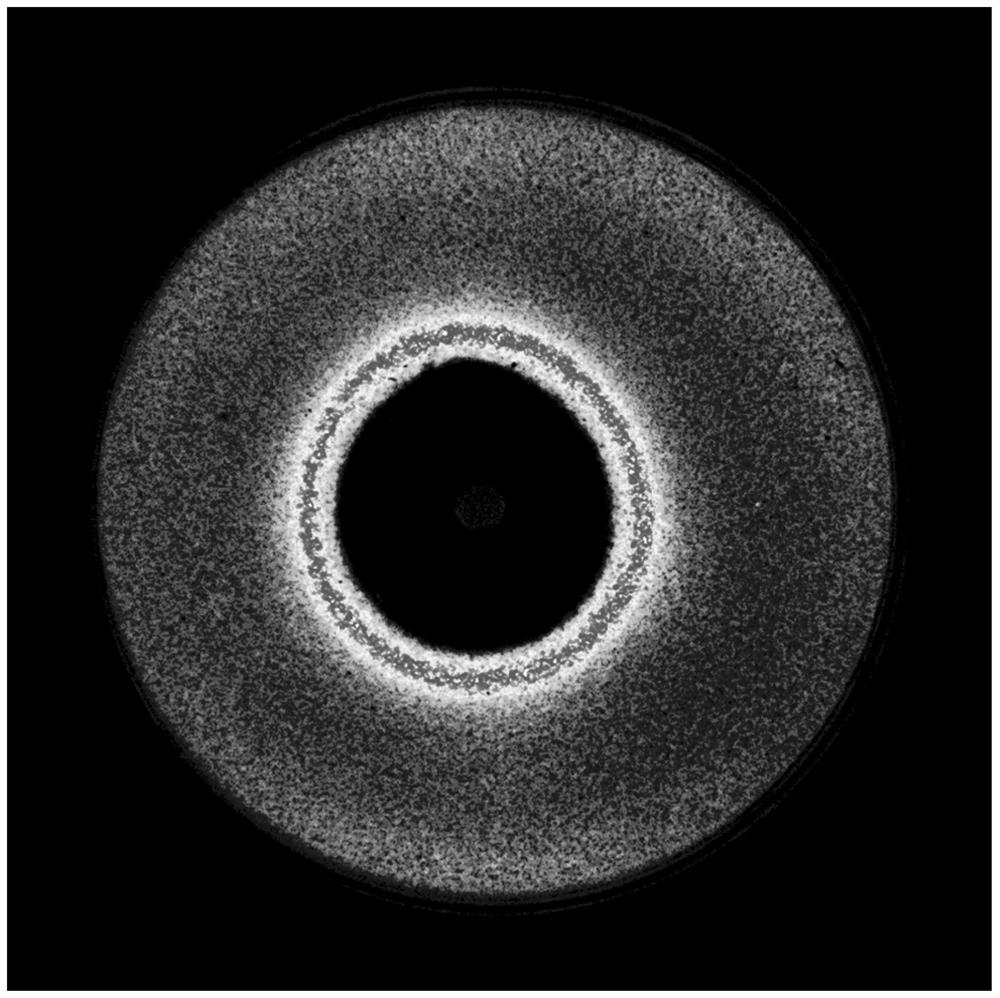
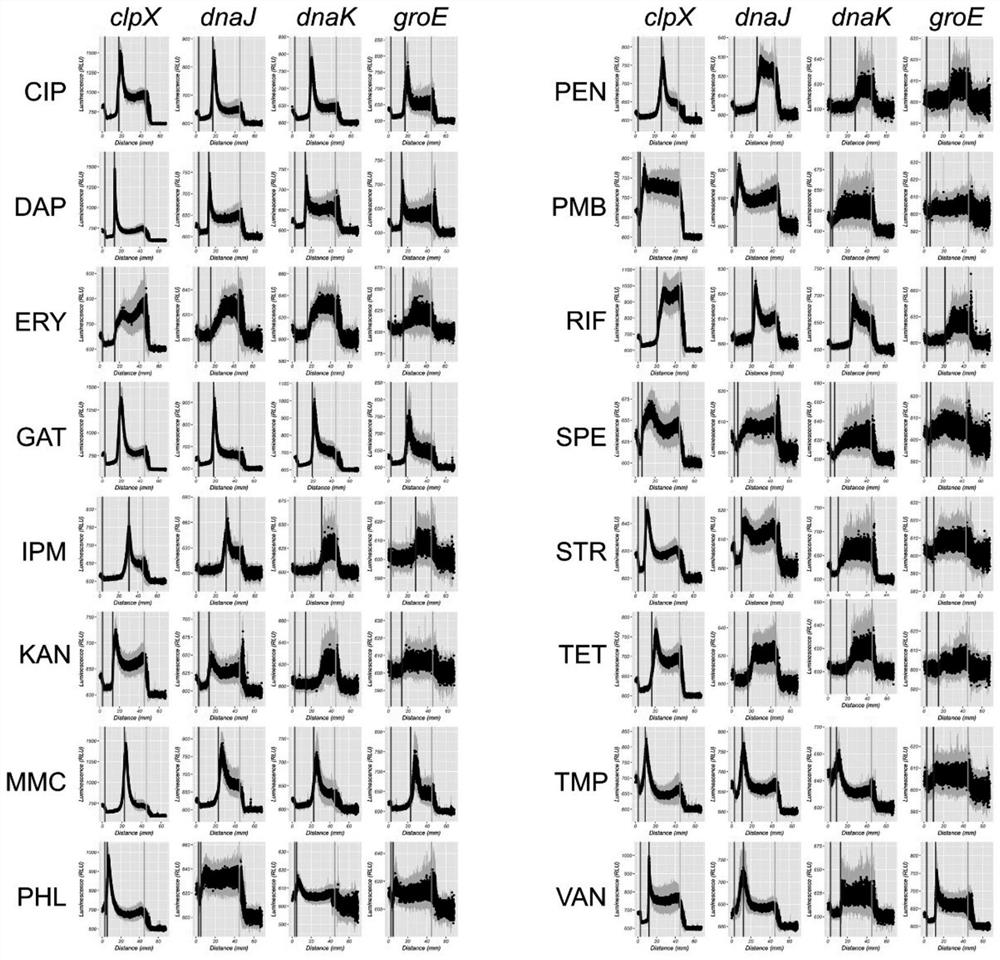
[0154] The drawing of embodiment 2 characteristic luminous curves
[0155] Experimental strains: The strains used in the experiment are four reporter strains of Staphylococcus aureus lux: S1, S2, S3, and S4.
[0156] Media required for experiment and its preparation
[0157] 1. NYE agar medium
[0158] 2. 0.7% agar: Weigh 0.7g of agar with a balance and add it to the Erlenmeyer flask, then add 100ml of deionized water, and sterilize under high pressure steam at 121°C for 20 minutes for later use.
[0159] Antibiotics required for experiments and their preparation
[0160] 1. Daptomycin (DAP) solution: Dissolve 30 mg of daptomycin in 1 ml of ultrapure water to make the final concentration 30 mg / ml, and filter through a 0.22 μm membrane to sterilize;
[0161] 2. Polymyxin B (PMB) solution: Dissolve 100mg of polymyxin B in 1ml of ultrapure water to a final concentration of 100mg / ml, and filter through a 0.22μm membrane to sterilize;
[0162] 3. Imipenem (IPM) solution: Dissol...
Embodiment 3
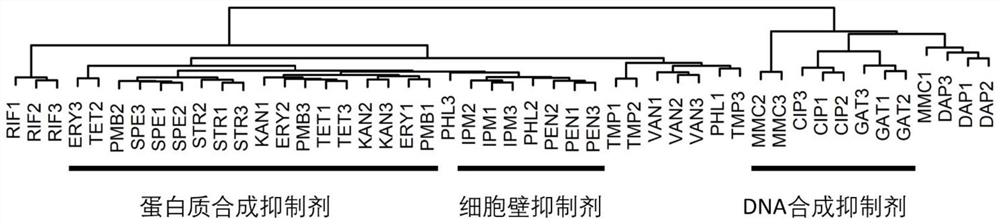
[0202] Cluster Analysis of Example 3 Characteristic Luminescence Curves
[0203] Experimental procedure
[0204] Step 1. Cluster analysis
[0205] 1. Record each characteristic luminous curve (from the edge of the inhibition zone to the edge of the plate) as a vector (luminous intensity characterization vector) represented by 300 luminous intensities, and each luminous intensity represents in the luminous picture of embodiment 2 The average luminous intensity in each circle from the edge of the inhibition zone to the edge of the plate (the span is 1 / 300 of the edge of the inhibition zone to the edge of the plate).
[0206] 2. Merge the characteristic luminescence curves of the four reporter strains corresponding to each antibiotic into a combined luminescence intensity characterization vector represented by 1200 luminescence intensities.
[0207] 3. Using the Euclidean distance calculation method, calculate the distance between the combined luminous intensity representation ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com