Multi-nucleic acid detection method and kit
A detection kit and detection method technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of long time consumption and insufficient sensitivity, and achieve the effect of short detection time, high sensitivity and multiple detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
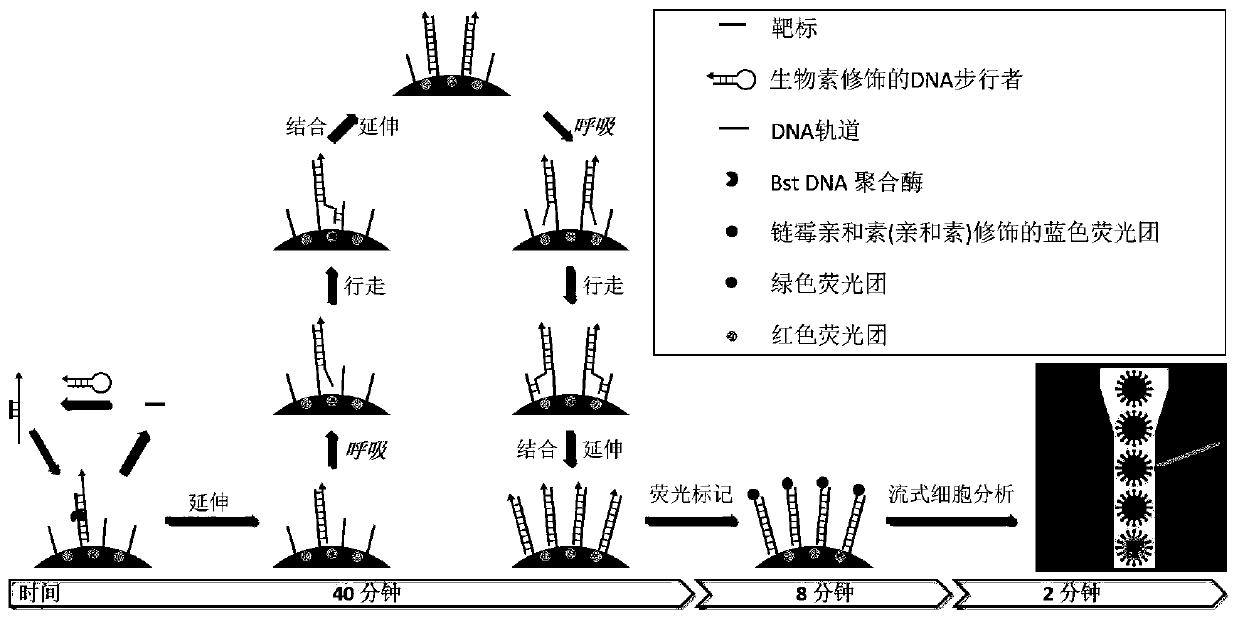
[0042] Nucleic acid sequence design of target miRNAs, DNA walkers and DNA tracks
[0043] For five targets (miRNA-29a, miRNA-151-5p, miRNA-15b, miRNA-26b, and miRNA-15a), DNA walkers (walker 1, walker 2) that can specifically hybridize to the corresponding target sequences were designed. , walker3, walker 4, and walker 5). The secondary structure of the DNA walker nucleic acid is a stem-loop structure. After forming a complementary double strand, the melting temperature of the end of the double strand is slightly lower than that of the middle region, enabling DNA respiration to occur. The DNA walker sequence consists of two parts: the target recognition region, which is used to hybridize with the target to open the stem-loop structure; the DNA track recognition region, which connects the DNA walker with the stem-loop structure opened and the DNA track (track 1, track 2, track 3 , track 4 or track 5) will be attached to the microsphere surface by hybridization. DNA track prov...
Embodiment 2
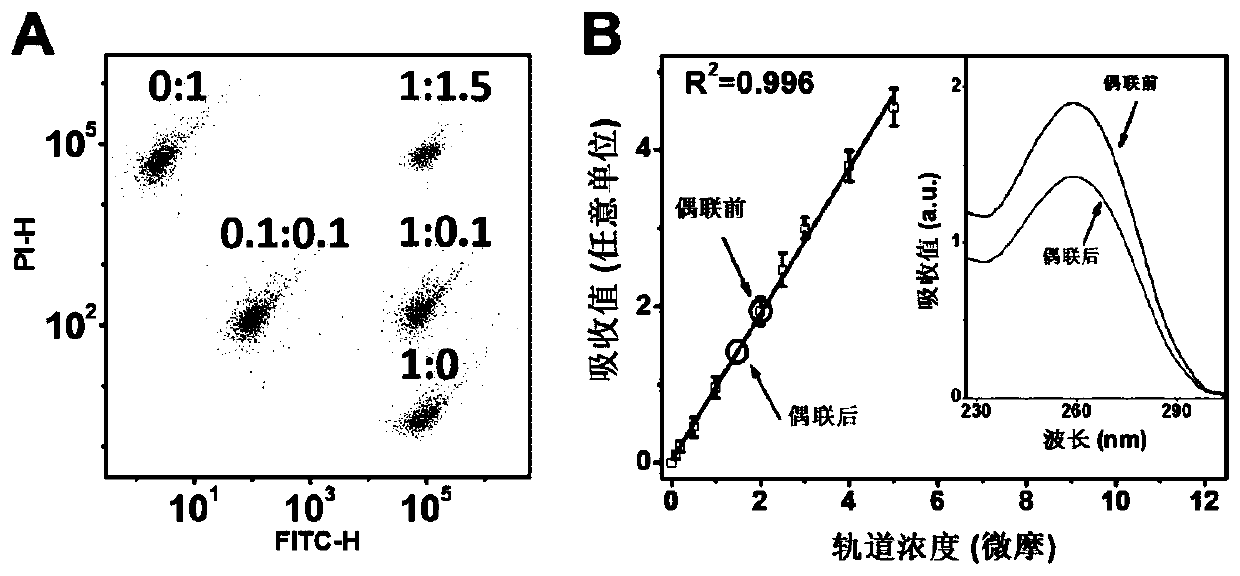
[0048] Preparation of fluorescently encoded microspheres loaded with DNA tracks
[0049] The green / red fluorescence intensity ratios of the five fluorescently encoded microspheres are 1:0, 0.1:0.1, 1:0.1, 1:1.5 and 0:1, respectively, such as figure 2 Shown in A, this figure shows flow cytometric characterization of five fluorescently encoded microspheres.
[0050] Add 50 mg / mL of EDC (1-(3-dimethylaminopropyl)- 3-Ethylcarbodiimide hydrochloride) and NHS (N-hydroxysuccinimide) solution, stirred for 30 minutes and then added 2 μl of 100 μM 5’ amino-modified DNA orbital. After stirring and reacting at room temperature for 3 hours, add 1 ml of phosphate buffer (pH=7.4) containing 0.1% Tween-20 by mass fraction, stir for 30 minutes and then centrifuge, the precipitated microspheres are dispersed in ultrapure water, and the loaded DNA track is obtained fluorescently encoded microspheres. Connect the five DNA tracks to the corresponding fluorescently encoded microspheres accordin...
Embodiment 3
[0055] DNA walking on the surface of fluorescently encoded microspheres
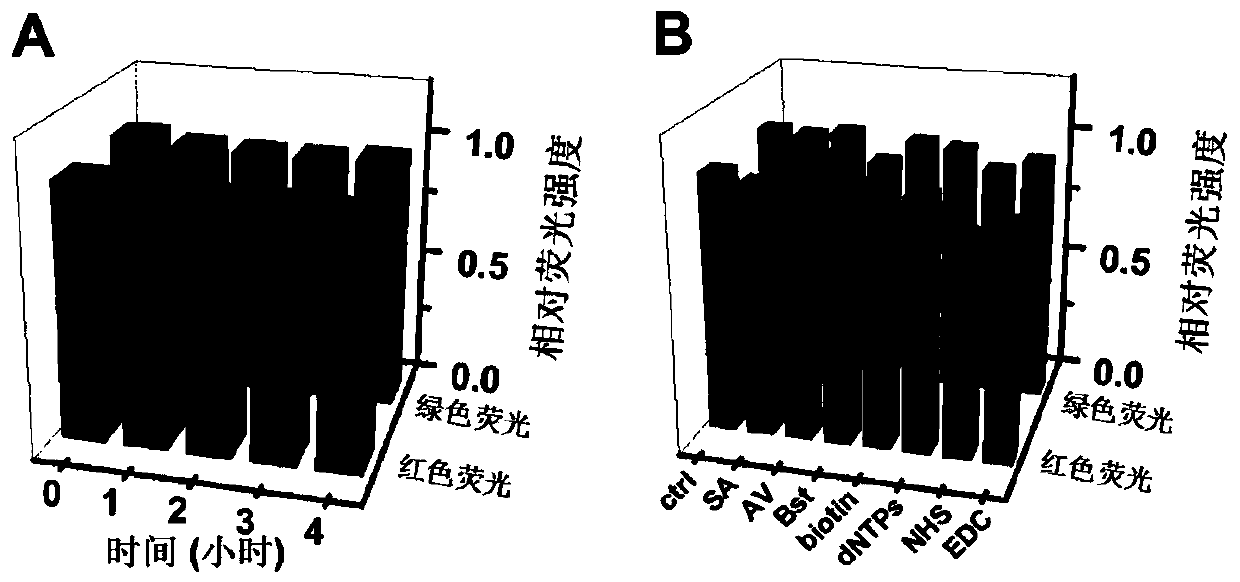
[0056] 2000 fluorescently encoded microspheres of the same loaded DNA track, 20 nM of the corresponding DNA walker, 100 fM of the corresponding miRNA, 2 μl of amplification buffer, 10 mM Mg in a 20 μl reaction volume 2+ , 10mM dNTPs and 1U / μL Bst DNA polymerase, and make up the remaining volume with water (amplification buffer, Mg 2+ and Bst DNA polymerase kit purchased from NEB Beijing, Cat. No. M0537S). 0, 10, 20, 30, 40, 50 and 60 minutes were selected for the reaction at 60°C, respectively. After centrifugation and washing, excess streptavidin-modified Pacific blue fluorescent dye (purchased from Thermo Fisher Scientific, item number S11222) was added, placed at 4 degrees Celsius after sufficient shaking, and centrifuged and washed after 30 minutes.
[0057] The result is as Figure 4 As shown, the results of flow cytometry show that miRNA-29a triggers the DNA walking reaction for 40 minutes, and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com