Preparation and application of low by-product pyridine formamide transformation microorganism
A picolinamide and microorganism technology is applied in the field of preparation and application of low-by-product picolinamide-converted microorganisms, and can solve the problems of high carboxypyridine content, affecting the use of picolinamide, increasing water, electricity, equipment, manpower, etc. The effect of the formation of carboxypyridine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1: Prepare low by-product pyridinecarboxamide transformed microorganisms as follows:
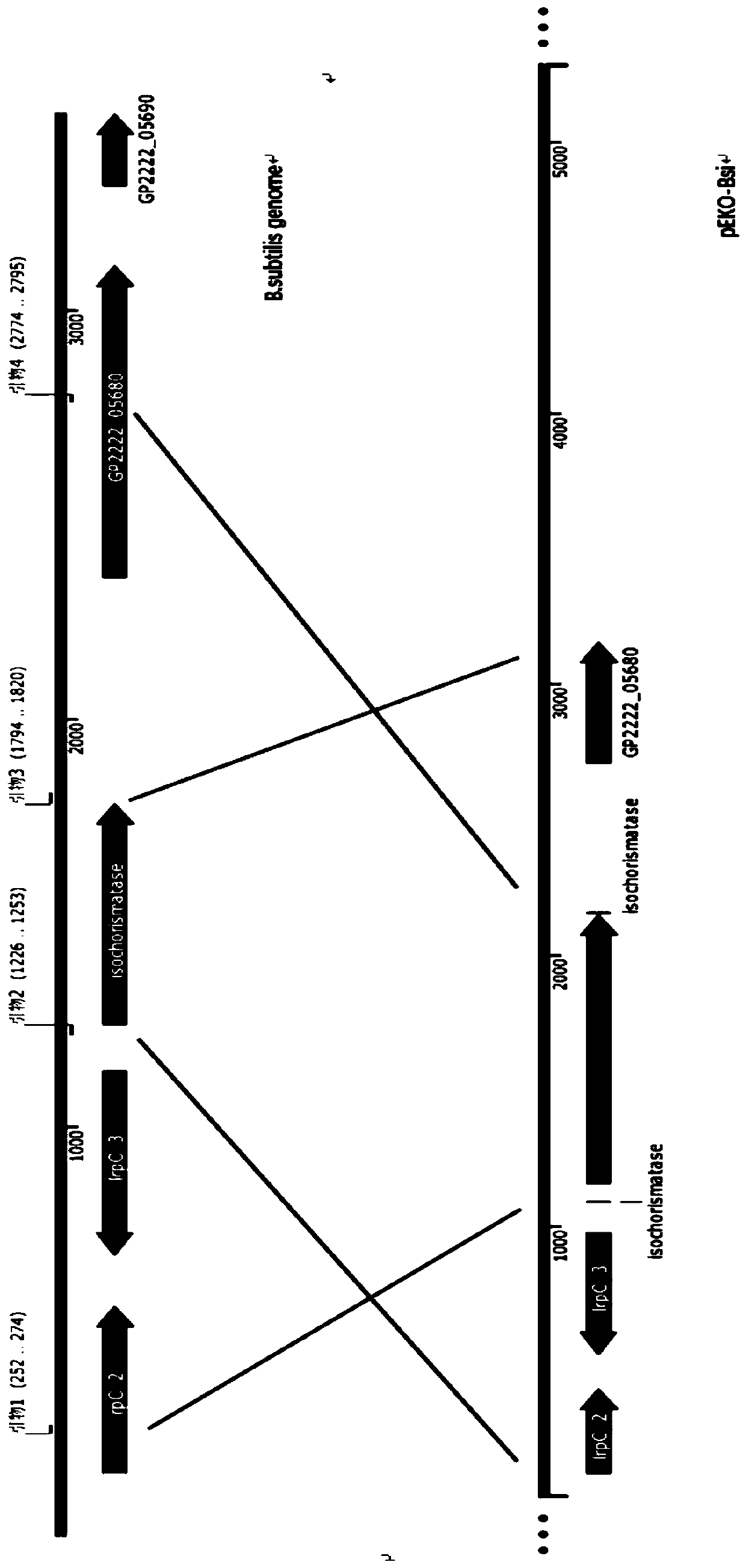
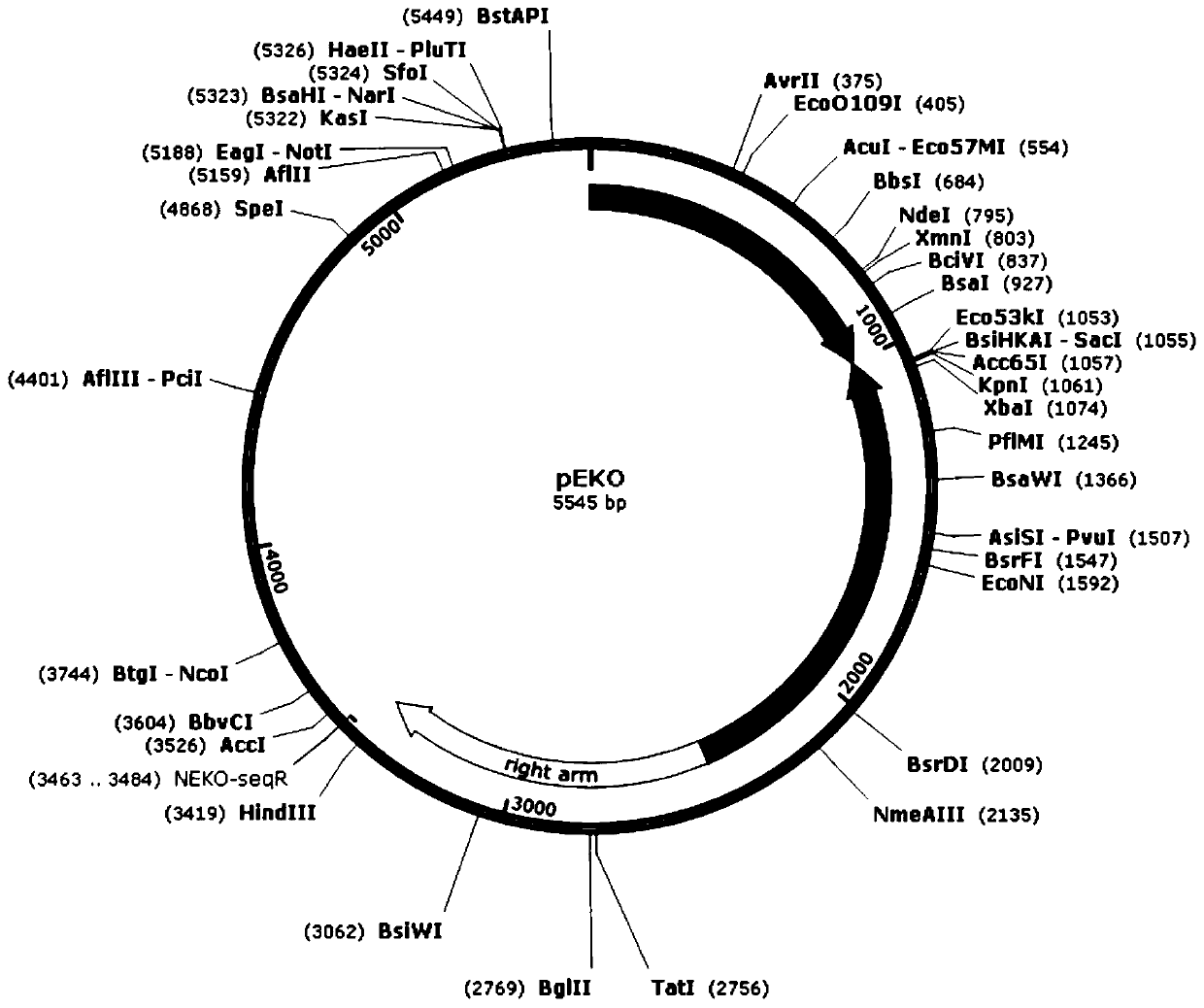
[0046] 1. Construction of gene knockout plasmid pEKO-Bsi
[0047] Mutation primers were designed with the isochorismatase (isochorismatase) sequence SEQ NO.1 of Bacillus subtilis as the target gene:
[0048] Primer 1: TAAAAAGGATCATCGGATCCCAGCAACCGCATCAAGAGTAGT
[0049] Primer 2: CGCGCAGGAAATTCTTTTTTTCACCTCTTAAAATTTTTATACT
[0050] Primer 3: GGTGAAAAAAAGAATTTCCTGCGCGAACATAACG
[0051] Primer 4: GACCATGATTACGCCAAGCTTTTGGGTGCTTGAATACTAATCC
[0052] SEQ NO.11 is the primary sequence of the protein corresponding to SEQ NO.1. The primer design must consider whether the change of amino acid residues in the primary sequence of the protein caused by gene mutation can lead to the loss or reduction of enzyme activity, and avoid the occurrence of amino acid residues. Equivalent substitutions or changes in non-enzyme-related amino acid residues.
[0053] Primer 1 and primer 2, prim...
Embodiment 2
[0088] Embodiment 2: prepare low by-product pyridinecarboxamide transformed microorganisms as follows:
[0089] 1. Construction of gene knockout plasmid pEKO-Bci
[0090] Mutation primers were designed with the isochorismatase (isochorismatase) sequence SEQ NO.2 of Bacillus cereus ATCC 14579 as the target gene:
[0091] Primer 1: CATGCCATATTCAAAACGATAAGATGG
[0092] Primer 2: TACATATTCACGAAAGCGTACGTCCACTCCTTAGA
[0093] Primer 3: ACGCATAATCATTCACAATAGTTTAAATGGC
[0094] Primer 4: ATAAGCACGCAAGAATTTTTAGCTCTCAAACAT
[0095] SEQ NO.12 is the primary protein sequence corresponding to SEQ NO.2.
[0096] Primer 1 and primer 2, primer 3 and primer 4 respectively take Bacillus cereus ATCC14579 genome sequence as template, carry out PCR amplification reaction, PCR reaction conditions are as follows:
[0097] Pre-denaturation at 95°C for 5min; denaturation at 94°C for 60s, annealing at 58.5°C for 42s, extension at 72°C for 90s, 35 cycles, and finally extension at 72°C for 10min. S...
Embodiment 3
[0125] Embodiment 3: Prepare low by-product pyridinecarboxamide transformed microorganisms as follows:
[0126] 1. Construction of gene knockout plasmid pEKO-Epn
[0127] Mutation primers were designed with the sequence SEQ NO.3 of purine-nucleotide phosphorylase (purine-nucleotide phosphorylase) of Escherichia coli (Escherichia coli K-12 MG1655) as the target gene:
[0128] Primer 1: AGAATTCCAGACTACACATTAATGCAGAAATGGGCGATTTCGCTG
[0129] Primer 2: GCTGACTTCGACATGGTGCGTATCGCAGATCGACGATACAATA
[0130] Primer 3: ATATCCGGCTACGTCGCTGCAGAATTTGGCGCGAA
[0131] Primer 4: TGGAATCCGTTCTGCTGGGCGATAAAGAGAAACTAGACGT
[0132] SEQ NO.13 is the primary protein sequence corresponding to SEQ NO.3.
[0133] Primer 1 and primer 2, primer 3 and primer 4 respectively take Escherichia coli (Escherichia coli K-12 MG1655) genome sequence as template, carry out PCR amplification reaction, PCR reaction condition is as follows:
[0134] Pre-denaturation at 95°C for 5min; denaturation at 94°C for 40...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com