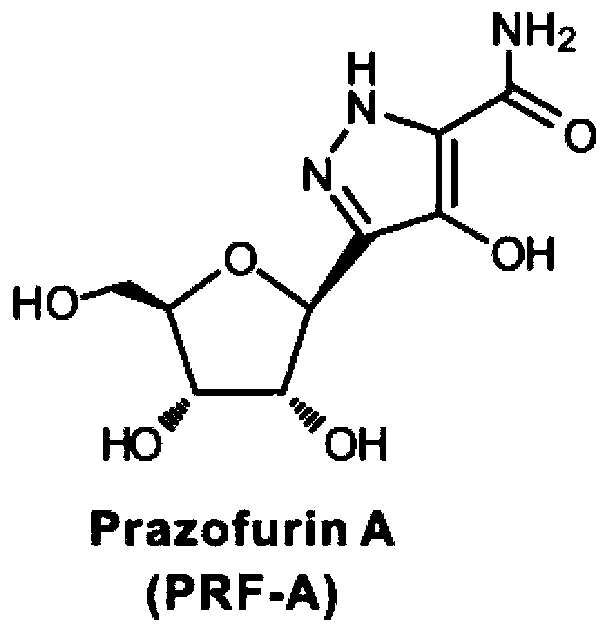
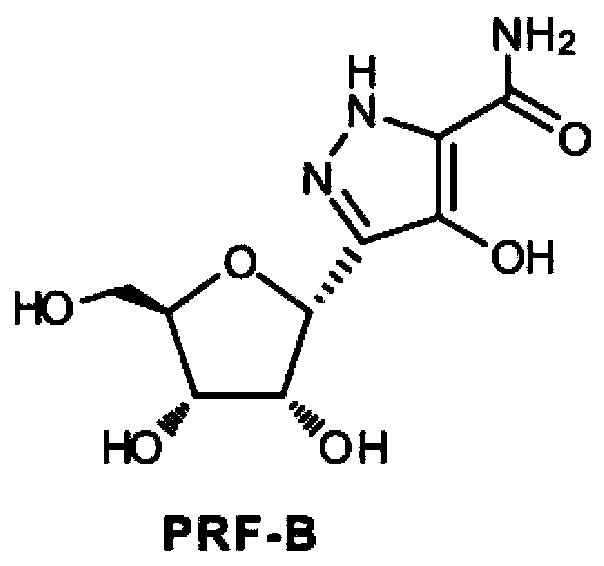
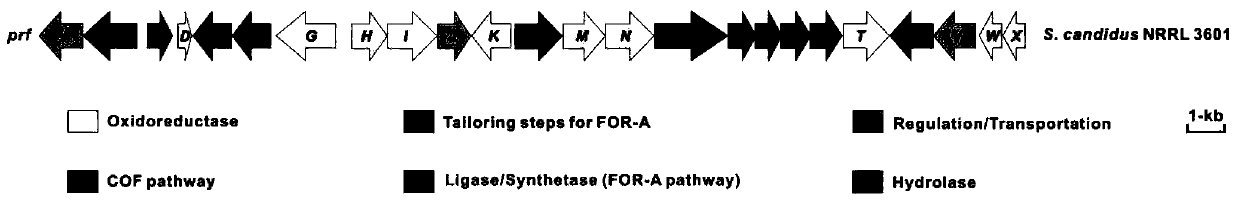
Pyrazomycin biosynthesis gene cluster, recombinant bacteria and application of recombinant bacteria
A pyrazomycin, biosynthesis technology, applied in the direction of plant genetic improvement, application, microorganisms, etc., can solve the problem of unclear biosynthesis mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] [Example 1] Extraction of total DNA of pyrazomycin-producing bacteria Streptomyces pure albicans NRRL 3601:
[0073] Take 30 μL of pure Streptomyces albicans NRRL 3601 spores into 50 mL of TSB medium, culture at 30°C, 200 rpm for 24 hours, until the medium becomes turbid. 50mL of pure Streptomyces albicans NRRL 3601 bacterial solution was centrifuged at 8,000rpm, 8°C for 10min to remove the supernatant and collect the bacteria. Dissolve the bacteria in 25mL of 10.3% sucrose solution, shake and mix well, wash the bacteria, centrifuge at 6,000rpm, 4°C, 10min to remove the supernatant; then dissolve the bacteria in 25mL of setbuffer, shake and mix, 6,000rpm, 4°C , centrifuged for 10 min to remove the supernatant, and repeated twice; dissolved the bacteria in 10 mL of set buffer, oscillated and mixed, added 50 μL of lysozyme solution (100 mg / mL) and placed it in a 37 ° C water bath for 30 min; then added 200 μL Proteinase K solution (50 mg / mL), after mixing, add 600 μL of ...
Embodiment 2
[0074] [Example 2] Fermentation conditions of pyrazomycin-producing bacteria Streptomyces pure albicans NRRL 3601, and product high-performance liquid phase (HPLC) detection conditions.
[0075]The spores of Streptomyces pure albicans NRRL 3601 were inoculated in the seed medium, cultured at 30°C, 220rmp for 48h, transferred to a fermentation shaker flask according to the inoculum size of 2%, and cultivated at 30°C, 220rmp for 7 days. Collect the fermentation broth, adjust the pH to 3-4 with oxalic acid, centrifuge the fermentation broth at 6,000 rpm for 20 minutes, remove the supernatant and pass through a 0.22 μm microporous membrane for HPLC detection and analysis.
[0076] HPLC detection conditions: phase A is ultrapure water added with 0.15% trifluoroacetic acid (TFA), and phase B is methanol. The initial 95% phase A gradient eluted to 25% within 30 minutes, and the A phase was converted to 50% at 41 minutes, and the A phase was converted to 95% at 42 minutes, and maintai...
Embodiment 3
[0077] [Example 3] The pyrazomycin-producing strain Streptomyces puree NRRL 3601 and its heterologous expression conjugative transfer method.
[0078] Transform the target plasmid to be conjugatively transferred into E. coli E.coli ET12567 / pUZ8002 competent, and verify after the transformant grows, inoculate the positive single clone in 5 mL of LB culture medium, cultivate overnight at 37°C, and the bacterial liquid Inoculate at 10% in 5mL LB medium and incubate at 37°C for 3-5h. Take the host Streptomyces spores and centrifuge at 4,500rpm for 3min, remove the supernatant, wash with ultrapure water twice, and centrifuge at 4,500rpm for 3min to remove the supernatant. Add 700 μL of TES, mix well, heat shock at 45°C for 10 minutes, then add 750 μL 2x spore pre-germination solution, and incubate at 30°C for 3-5 hours.
[0079] Centrifuge the cultured Escherichia coli at 4°C and 4,000rpm for 3 minutes, remove the supernatant and add 10mL LB to wash twice, then centrifuge to remov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com