Optimal ion pair selection method for quasi-targeted metabonomics analysis
A technology of metabolomics and ion pairing, applied in the direction of material separation, analytical materials, scientific instruments, etc., can solve problems such as unfavorable and ion pair optimization, achieve the effect of improving coverage, improving work efficiency, and facilitating subsequent data processing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
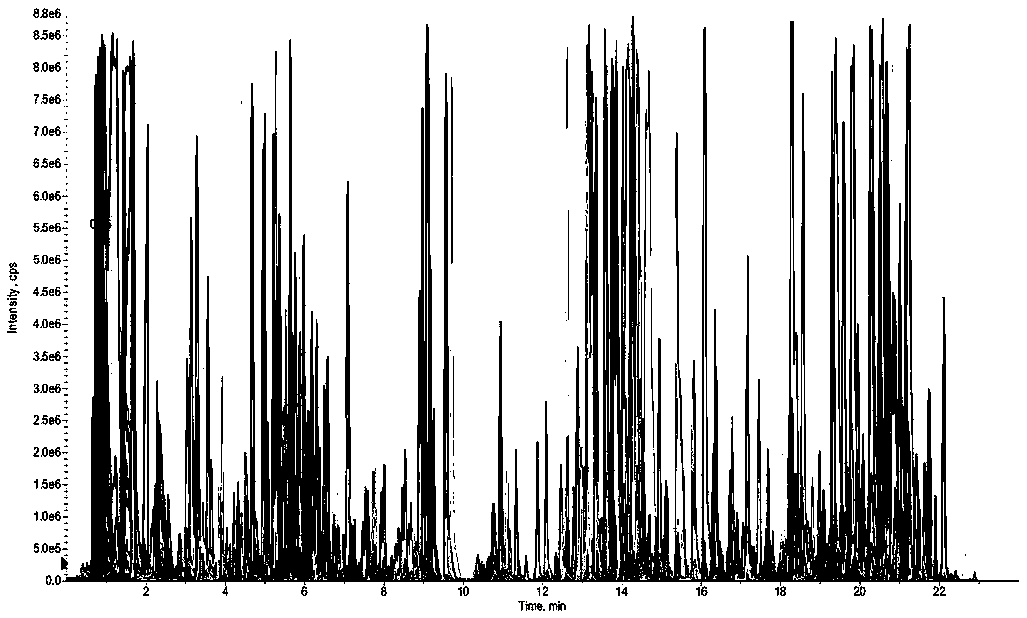
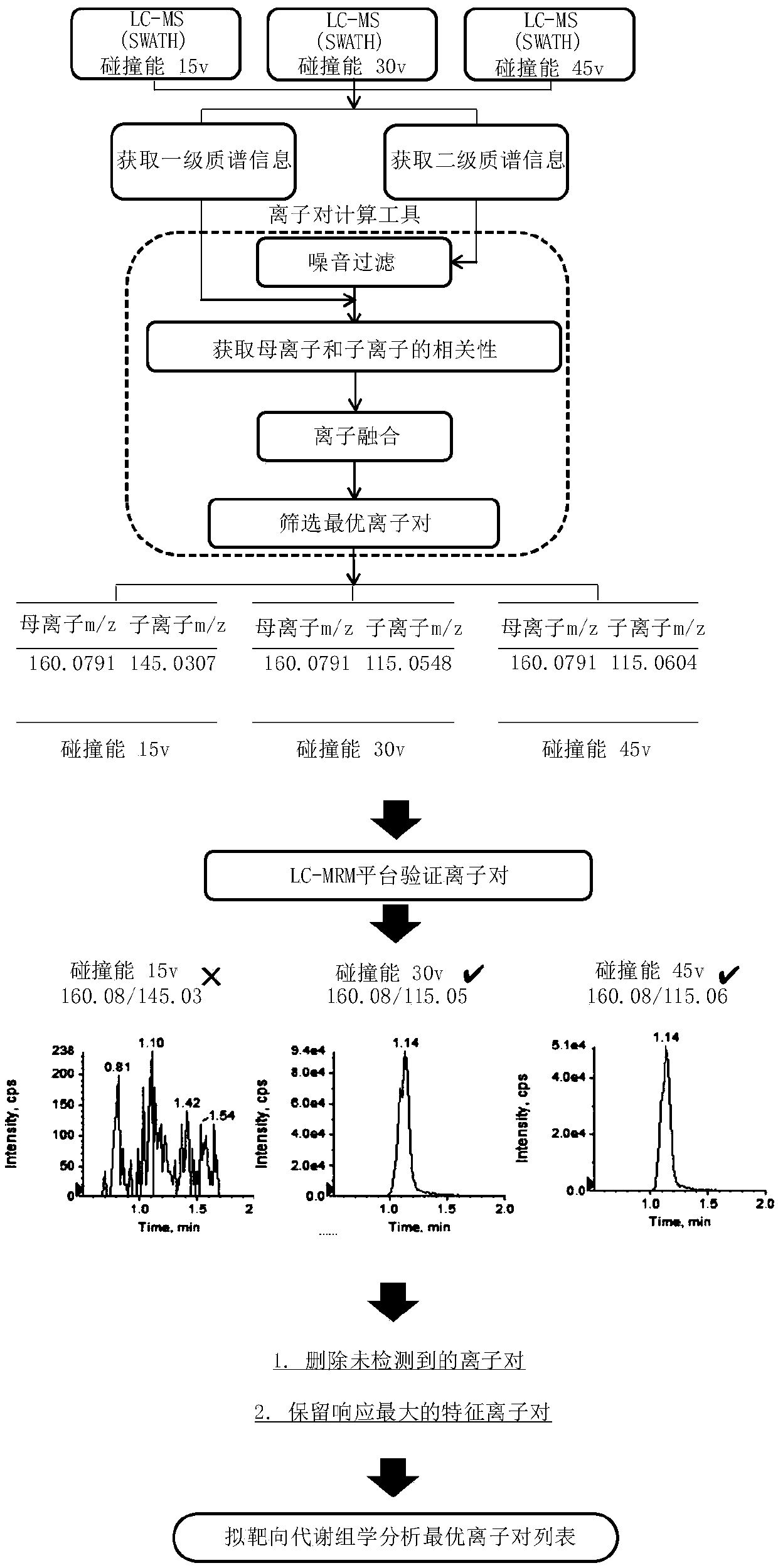
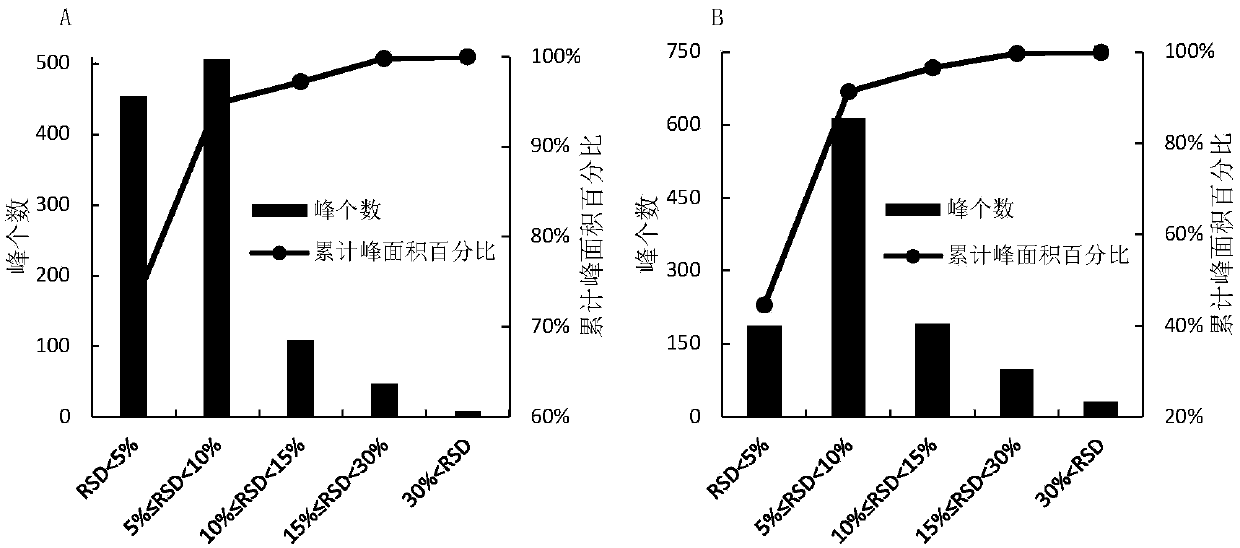
[0045] Establishment of a quasi-targeted metabolomics analysis method for serum samples based on mass spectrometry information-independent acquisition mode:
[0046] 1) Take 200 μL of 10 healthy human serum samples and mix them as quality control samples. Take 100 μL of quality control samples in a 1.5mL ep tube and add 400 μL of methanol solvent to vortex to remove protein and extract metabolites, vortex for 30s After fully mixing the solvent and the sample, the sample was immediately placed in a centrifuge, and centrifuged at 4° C. and 14,000 rpm for 15 min. After centrifugation, 400 μL of the supernatant was drawn into a 1.5 mL ep tube for low-temperature centrifugation and freeze-drying. Finally, 50 μL of 20% acetonitrile water (acetonitrile: water = 4:1, V / V) was added to the freeze-dried sample for reconstitution, centrifuged at 4°C and 14,000 rpm for 10 min, and the supernatant was absorbed for subsequent liquid Phase chromatography-mass spectrometry detection and anal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com