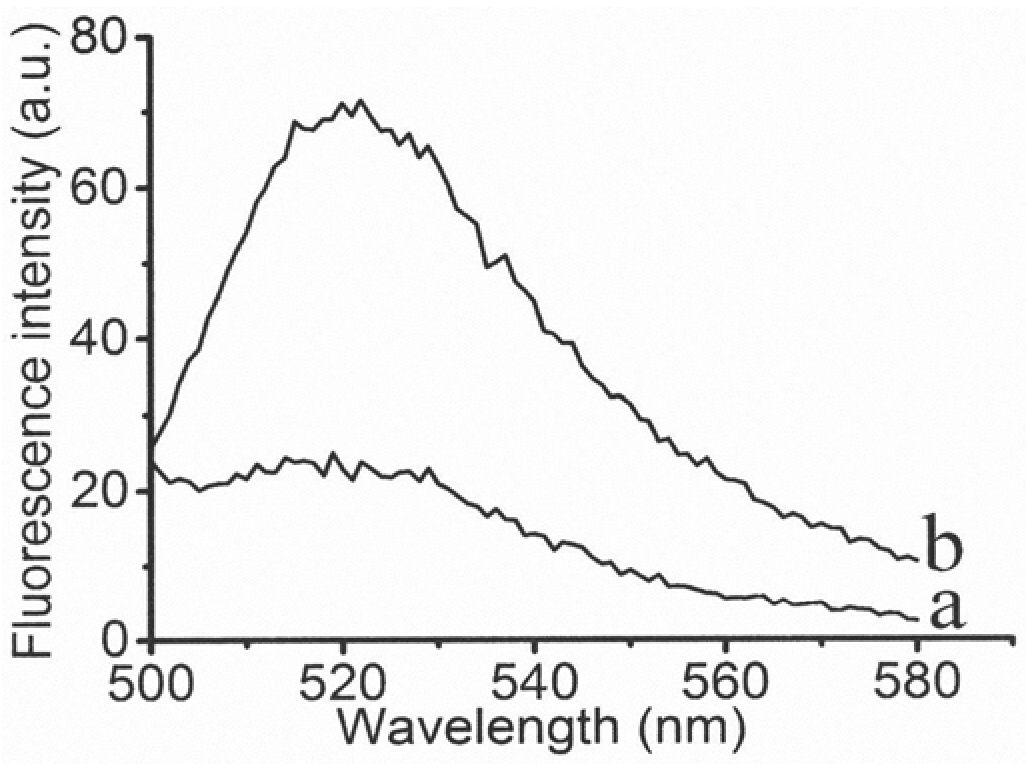
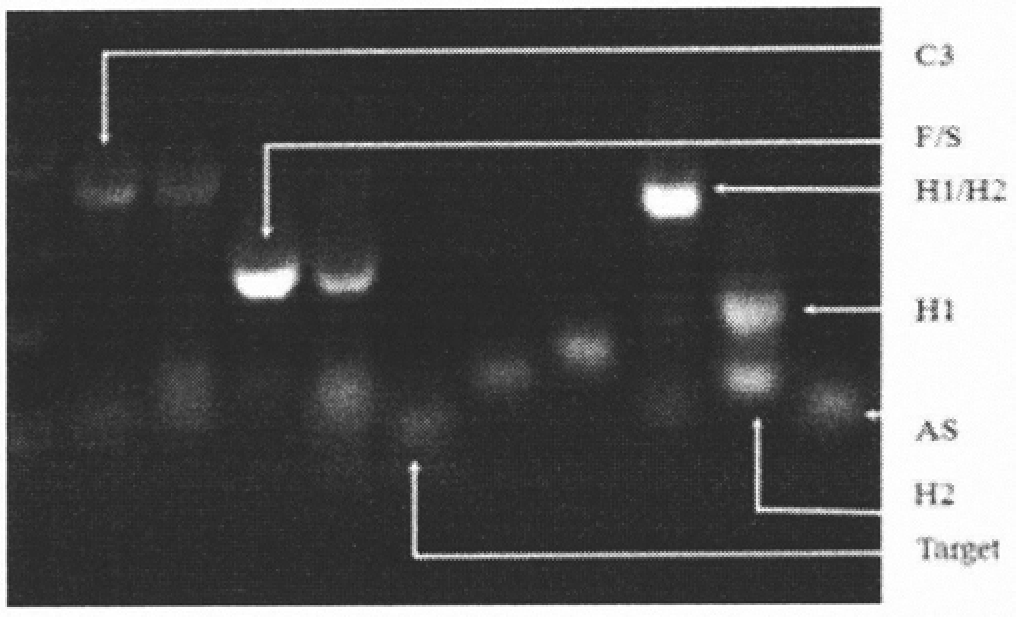
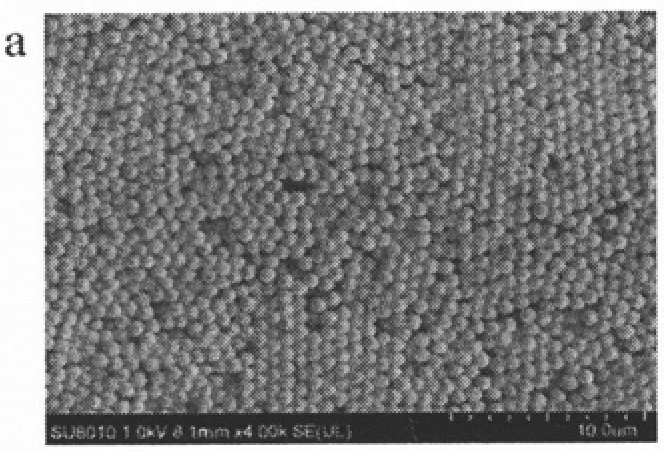
MicroRNA biosensor coupling 3D DNA walking machine with catalytic hairpin assembly
A biosensor, 3-DDNA technology, applied in the field of biosensors, can solve the problems of reducing the colloidal stability, hindering DNA hybridization, and the connecting force is not strong enough.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0043] To prepare a biosensor based on entropy-driven 3-D DNA walking machine coupled with catalytic hairpin self-assembly, follow the steps below:
[0044] (1) Preparation of 3-D DNA walking machine
[0045] The purified three nucleic acid strands LS, AS, S and TEmg buffer are annealed and then refolded to prepare the LS / AS / S triple nucleic acid complex (C3). The ratio of LS:AS:S is 1:1.5:1.5. The annealing process is to heat the solution in a PCR machine to 95°C for 5 minutes, and then slowly lower the temperature to 25°C at a rate of 0.1°C per second. Then, 50 μL of streptavidin-modified polystyrene microspheres (PS) were centrifuged at 12,000 rpm for 5 minutes, washed twice with binding buffer, and centrifuged to discard the supernatant. Resuspend in 50μL binding buffer and add 5μLμM C3. After incubating for 15 minutes in a 37°C incubator, wash with TEMg buffer 3 times, centrifuge at 12,000 rpm for 5 minutes to discard the supernatant. Finally, resuspend with 50μL of TEMg b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com