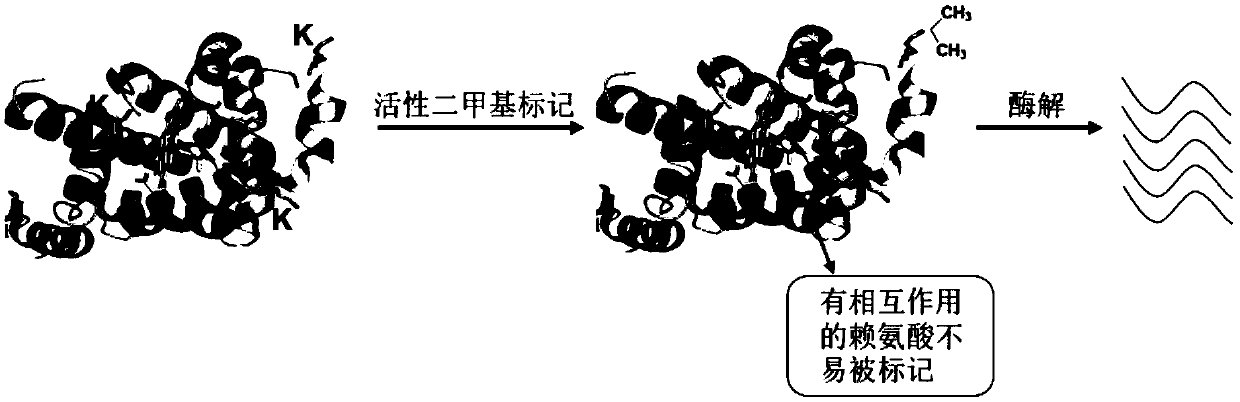
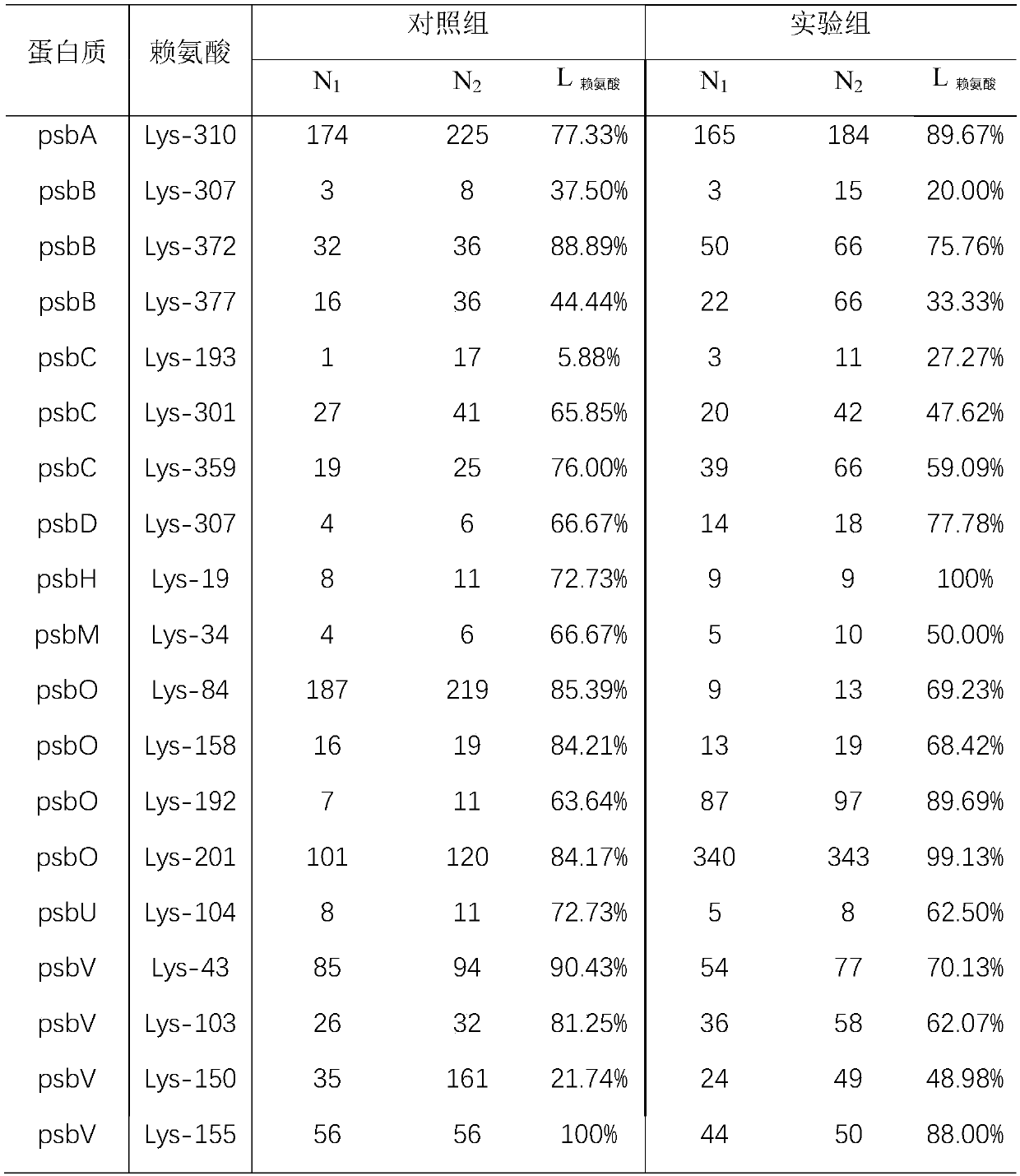
Chemical labeling and mass spectrometry-based structure analysis method for protein and material interaction
A technology of chemical labeling and analysis method, applied in the field of protein-material interaction structure analysis based on chemical labeling and mass spectrometry, can solve the problems of protein structure change, affecting protein electrostatic interaction, loss of activity, etc. Effects of structure and activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Structural analysis of protein-material interaction based on mass spectrometry:
[0053] 1. Prepare two groups of photosystem II proteins with a concentration of 0.01 μg / μL for the control group and the experimental group. Add iron oxide to the protein and materials in the experimental group at a ratio of 1:100, and incubate for 10 minutes at the same time;
[0054] 2. Add formaldehyde with a final concentration of 0.01mmol / L and sodium cyanoborohydride with a final concentration of 0.01mmol / L respectively, and react at 4°C for 120min;
[0055] 3. Add NH with a final concentration of 1mmol / L 4 HCO 3 ;
[0056] 4. Replace the solution obtained in the above step 3 with 8M urea, boil it, add chymotrypsin, the mass ratio of protein to enzyme is 50:1, and enzymolyze overnight at 25°C;
[0057] 5. The solution obtained in the above step 4 is acidified, and the chromatographic separation and the detection of mass spectrometry are carried out;
[0058] 6. Perform a database...
Embodiment 2
[0065] Structural analysis of protein-material interaction based on mass spectrometry:
[0066] 1. Prepare two groups of photosystem II proteins with a concentration of 0.1 μg / μL for the control group and the experimental group. Add iron oxide to the protein and materials in the experimental group at a ratio of 1:20, and incubate for 30 minutes at the same time;
[0067] 2. Add formaldehyde with a final concentration of 2mmol / L and sodium cyanoborohydride with a final concentration of 2mmol / L respectively, and react for 30min at room temperature;
[0068] 3. Add CH with a final concentration of 50mmol / L 3 COONH 4 ;
[0069] 4. Replace the solution obtained in the above step 3 with 6M guanidine hydrochloride, boil it, add chymotrypsin, the mass ratio of protein to enzyme is 25:1, and enzymolyze at 30°C overnight;
[0070] 5. The solution obtained in the above step 4 is acidified, and the chromatographic separation and the detection of mass spectrometry are carried out;
[0...
Embodiment 3
[0078] Structural analysis of protein-material interaction based on mass spectrometry:
[0079] 1. Prepare two groups of photosystem II proteins with a concentration of 10 μg / μL for the control group and the experimental group. Add iron oxide to the protein and materials in the experimental group at a ratio of 1:10, and incubate for 60 minutes at the same time;
[0080] 2. Add formaldehyde with a final concentration of 100mmol / L and sodium cyanoborohydride with a final concentration of 100mmol / L respectively, and react at 40°C for 5 minutes;
[0081] 3. Add NH with a final concentration of 1mmol / L 4 HCO 3 ;
[0082] 4. Replace the solution obtained in the above step 3 with 6M urea, boil it, add trypsin, the mass ratio of protein to enzyme is 25:1, and enzymolyze overnight at 37°C;
[0083] 5. The solution obtained in the above step 4 is acidified, and the chromatographic separation and the detection of mass spectrometry are carried out;
[0084] 6. Perform a database searc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com