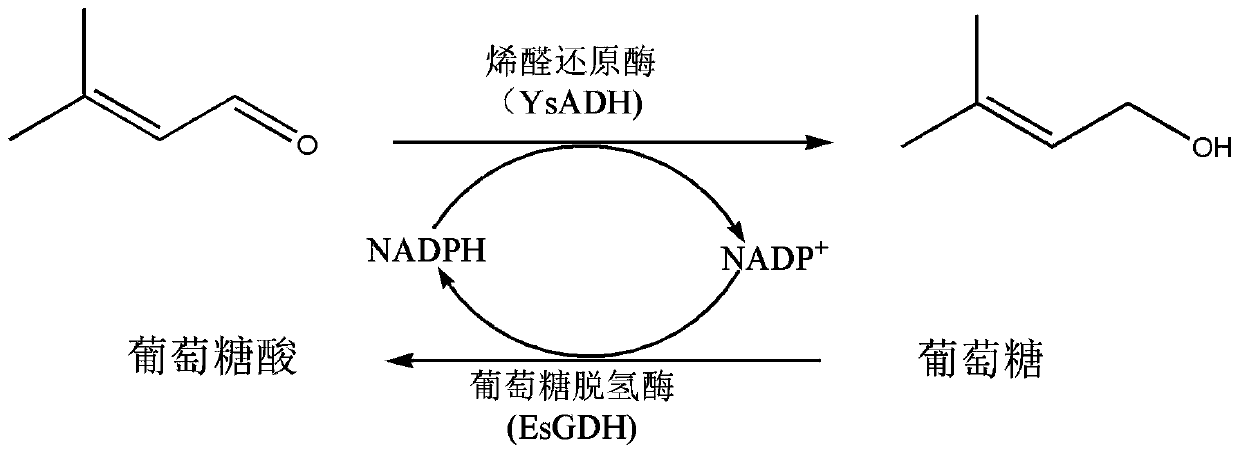
Recombinant genetically engineered bacterium of co-expressing olefine aldehyde reductase and glucose dehydrogenase and application of recombinant genetically engineered bacterium
A technology of glucose dehydrogenase and genetically engineered bacteria, applied in genetic engineering, oxidoreductase, applications, etc., can solve the problems that have not yet been seen in co-expressing enaldehyde reductase and glucose dehydrogenase recombinant E. Large-scale industrial production, lower production cost, high vitality effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Acquisition of the gene encoding alkenal reductase of Yokenella sp. WZY002
[0040] Using the published alkenal reductase encoding gene (GenBank accession number KF887947) derived from Yokenella sp. WZY002, after codon optimization, artificial synthesis (Suzhou Jinweizhi Biotechnology Co., Ltd. provides gene synthesis services ) The alkenal reductase encoding gene, the amino acid sequence and nucleotide sequence are shown in SEQ ID NO.1 and SEQ ID NO.2 respectively.
Embodiment 2
[0041] Example 2 Acquisition of D-glucose dehydrogenase encoding gene derived from Exiguobacterium
[0042] Using the published gene encoding D-glucose dehydrogenase from Exiguobacterium sibiricum (GenBank accession number ACB59697.1), after codon optimization, artificial synthesis (Suzhou Jinweizhi Biotechnology Co., Ltd. provides gene synthesis service) The gene encoding D-glucose dehydrogenase derived from Exiguobacterium, the amino acid sequence and nucleotide sequence are shown in SEQ ID NO.3 and SEQ ID NO.4, respectively.
Embodiment 3
[0043] Example 3 Construction of recombinant genetically engineered bacteria co-expressing alkenaldehyde reductase and D-glucose dehydrogenase
[0044] The alkenol reductase encoding gene (SEQ ID NO. 2) and the D-glucose dehydrogenase encoding gene (SEQ ID NO. 4) were inserted into Nco I, Hind III and Nde I, Xho on the pACYCDuet1 vector by "one-step cloning" Between two pairs of restriction sites, the recombinant plasmid pACYCDuet-1-YsADH-EsGDH ( figure 2 ). The recombinant plasmid pACYCDuet-1-YsADH-EsGDH was transformed into E.coli BL21(DE3) to obtain genetically engineered bacteria E.coli BL21(DE3) / pACYCDuet-1-YsADH-EsGDH.
[0045] The recombinant genetically engineered bacteria E.coliBL21(DE3) / pACYCDuet-1-YsADH-EsGDH was inoculated on LB solid medium containing 50 μg / mL chloramphenicol and streaked, and a single colony was picked and inoculated with 50 mL LB liquid medium. and add the final concentration of 50 μg / mL chloramphenicol, culture at 37 °C and 200 rpm constant ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com