Pathogenic factor for negatively regulating ustilaginoidea virens spore production, gene and application thereof
A technology of rice smut bacteria and pathogenic factors, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of rarely reported pathogenic factors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Identification and knockout of UvPSR1 gene in rice false smut.
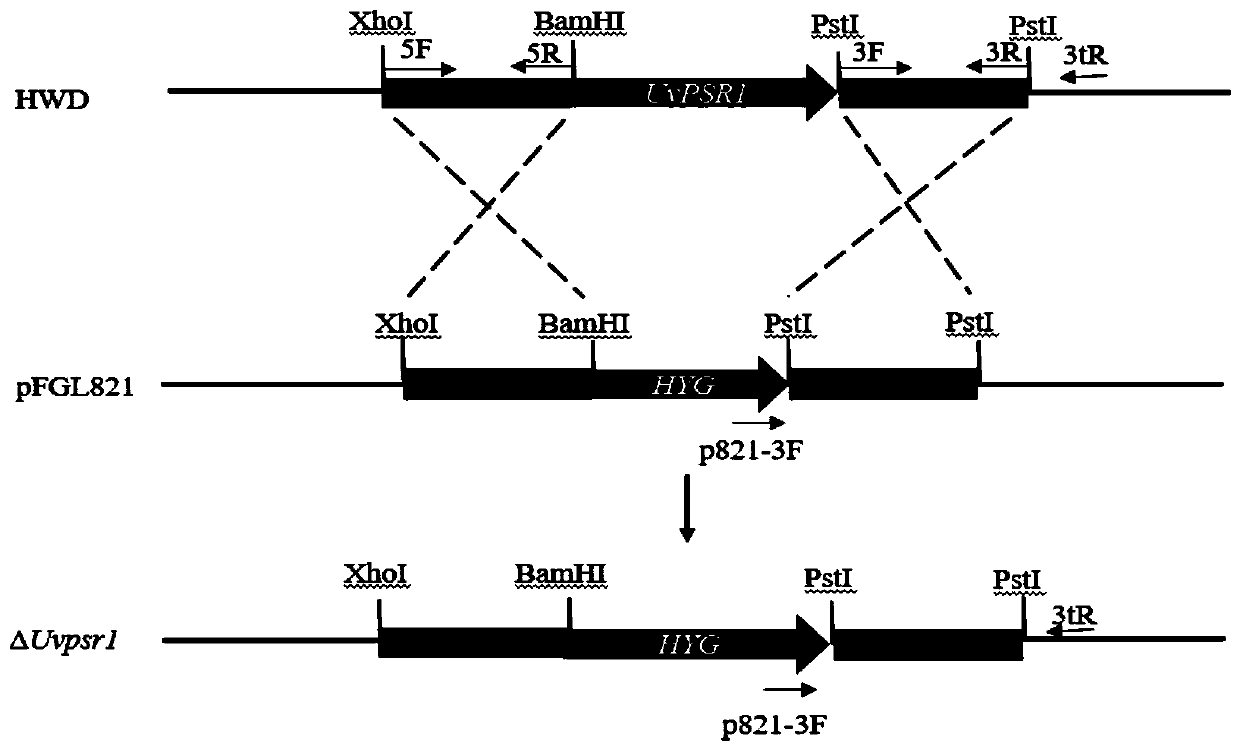
[0027] The amino acid sequence of rice smut UvPSR1 protein is shown in SEQ ID No.1, and the gene sequence (CDS sequence) of rice smut UvPSR1 gene is shown in SEQ ID No.2. The rice false smut UvPSR1 gene knockout vector was constructed by homologous recombination strategy. The knockout strategy is shown as figure 1 shown.
[0028] Knockout vector construction was performed as follows:
[0029] The UvPSR1 gene is 3556bp in length (sequence shown in SEQ ID No.3, including CDS sequence and some non-coding region sequences), which was replaced by the hygromycin B gene by homologous recombination to obtain the UvPSR1 knockout mutant . Specific steps are as follows:
[0030] Using the genome of the wild strain HWD of rice mistletoe (from Huang Junbin's laboratory of Huazhong Agricultural University) as a template, PCR amplified the upstream 1069bp of the UvPSR1 gene (the gene fragment is named UvPSR1-5') and...
Embodiment 2
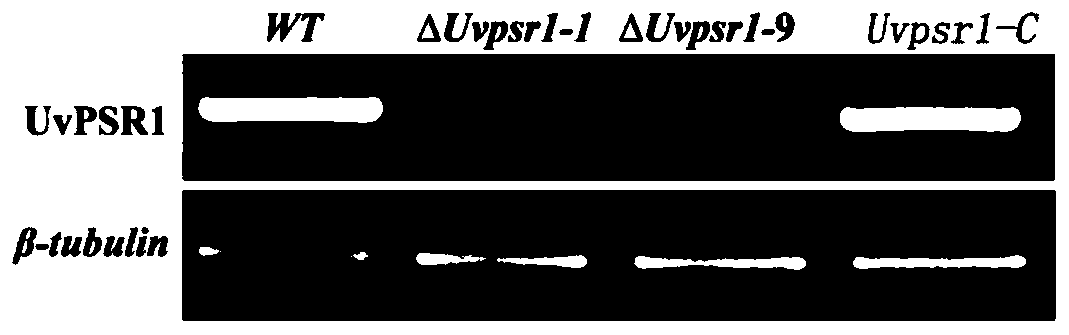
[0044] Identification of UvPSR1 gene knockout mutants of rice smut.
[0045] 1. Genomic DNA level identification
[0046] Extract the transformant genome, use the verified primers Uvpsr1-3tR (5'-CGGGCTCACTCCTGATTCTT-3'), and p821-5F (GTGGTGTAAACAAATTGACGCTTAG) to amplify the recombined fragment, and the detection system is:
[0047]
[0048] The reaction conditions are:
[0049] Pre-denaturation at 95°C for 30s;
[0050] Denaturation at 95°C for 7s, annealing at 63°C for 20s, extension at 72°C, the extension time was determined according to the size of the amplified fragment (30s / kb), a total of 34 cycles, and the annealing temperature decreased by 0.2°C for each cycle;
[0051] Extend at 72°C for 2 min.
[0052] If the amplification result is positive, it is a knockout mutant. A total of 7 mutant strains were identified, and the knockout mutants were named ΔUvpsr1-1, 2, 3, 7, 8, 9, and 13. Since the phenotypes of the knockout mutant transformants were consistent, ΔUvpsr1...
Embodiment 3
[0061] Acquisition of UvPSR1 deletion mutant complementation vector and complementation strain.
[0062] In order to verify that the phenotype of the mutant is caused by the deletion of UvPSR1, we constructed the complementary vector UvPSR1-C, using the wild-type HWD genome as a template, and using primers UvPSR1-F / R to amplify the full length of the UvPSR1 gene, using T4 Ligase ligated to pFGL820 (addgene ID: 58221) vector linearized with endonucleases EcoR1 and Xba1.
[0063] UvPSR1-F: 5'-CGGAATTCCGTCGGAGGCAACATTCAAG-3';
[0064] UvPSR1-R: 5'-GCTCTAGATCCTCGCAAATCCCCACTCT-3'.
[0065] The completed complementation vector was transformed into the knockout mutant ΔUvpsr1-1 strain, which was named Uvpsr1-C. QRT-PCR detection showed that UvPSR1 gene was normally expressed in the complementation mutant.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com