A Quantitative Analysis Method Integrating Proteome and Glycoproteome
A proteome and quantitative analysis technology, applied in the field of biochemistry, can solve the problems of lack of integrated proteome and glycoproteome quantitative analysis methods, loss of specific information of glycosylation sites, and low abundance of glycosylated peptides. , to achieve the effect of simple and time-saving steps, reliable quantitative results, and avoiding the influence of sialic acid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
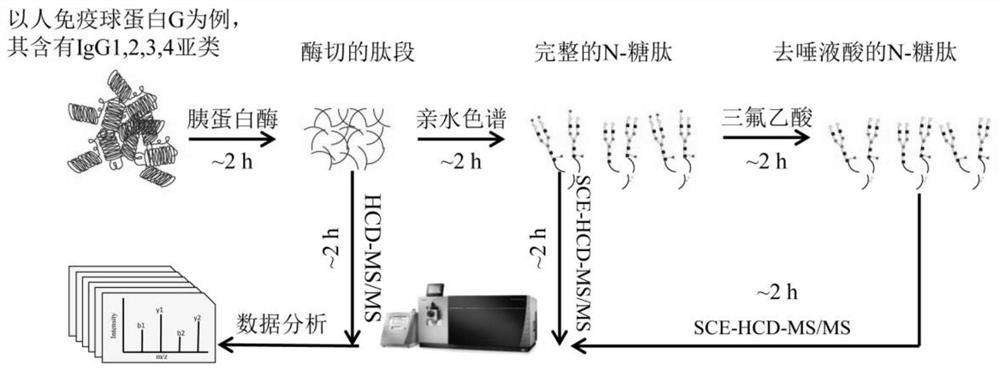
[0058] Example 1 Preparation of proteomic and glycoproteomic samples of protein IgG
[0059] 1. Method
[0060] (1) Dissolve 100 μg of commercial human IgG (Sigma) in 200 μL of 50 mmol / L ABC solution, and heat at 95° C. for 10 min for denaturation.
[0061] (2) DTT with a final concentration of 20 mmol / L was added to react at 56°C for 45 min, and IAM with a final concentration of 50 mmol / L was added to react for 1 h at room temperature in the dark.
[0062] (3) All the reaction solution was added into a 10KD ultrafiltration tube, centrifuged at 13000g for 15min×3 times, and washed with 200μL of 50mmol / L ABC each time.
[0063] (4) Add 100 μL of 50 mmol / L ABC and 2 g of trypsin (Promega), continue the reaction in a shaker at 37 °C for 2 h, centrifuge at 13,000 g for 15 min × 3 times, collect the peptide fragments, take 1 μg for storage, and the rest after lyophilization- Reserve at 80°C.
[0064] (5) Weigh 5 mg of Venusil HILIC filler (Bonna Aijer), rinse the filler with 0.1...
Embodiment 2
[0074] Example 2 Proteome and Glycoproteome Mass Spectrometry Analysis and Data Processing of Protein IgG
[0075] 1. Method
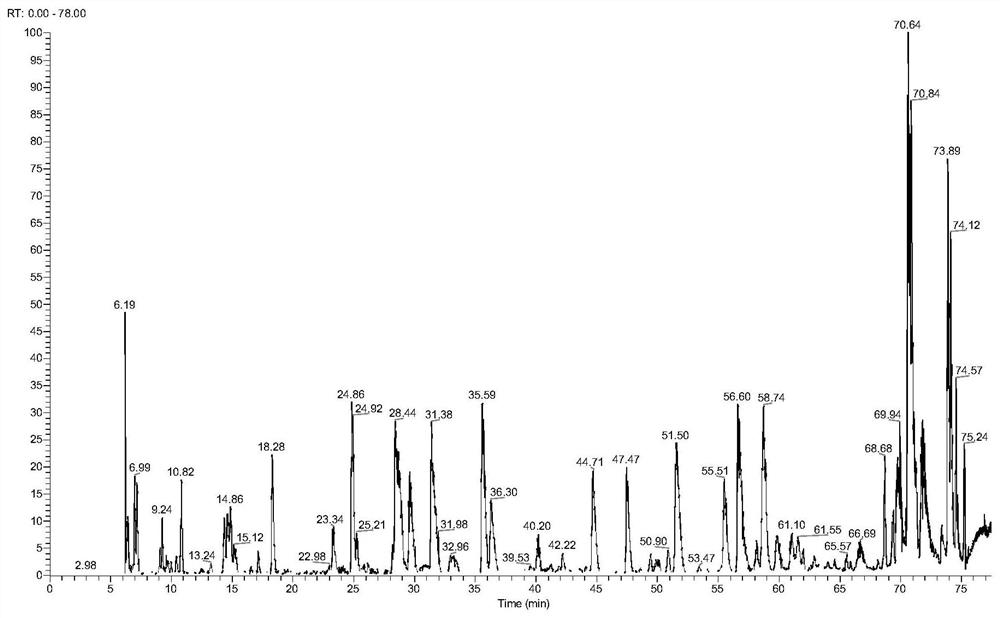
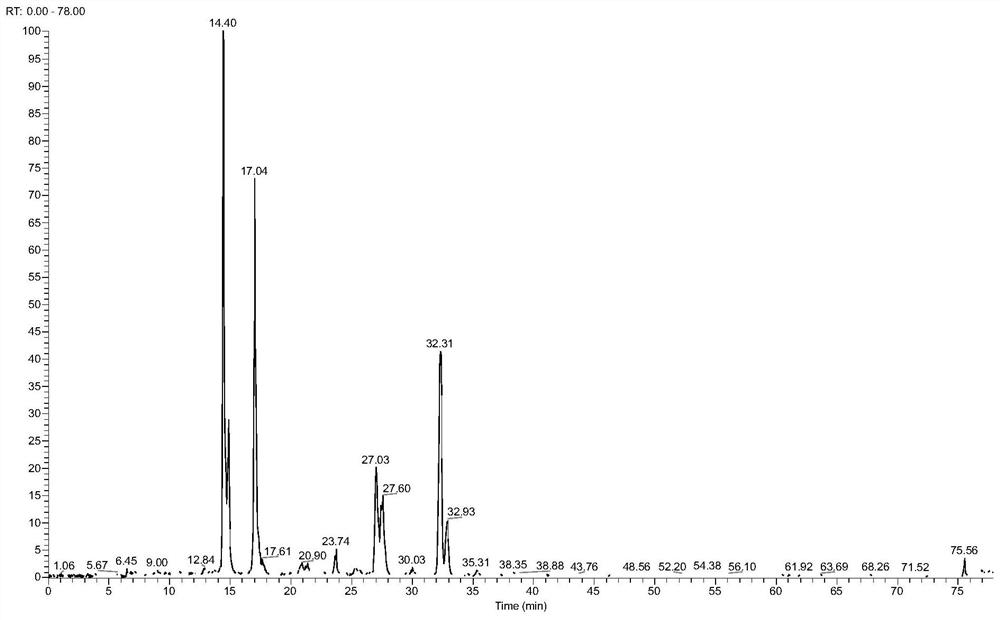
[0076] (1) Take 2 μL of the peptides, intact N-glycopeptides, and asialo-N-glycopeptides from steps (4) (8) (9) in Example 1 for mass spectrometry analysis. The instrument was an Orbitrap Fusion Lumos mass spectrometer.
[0077] (2) The chromatographic conditions are: chromatographic column: The inner diameter of the Magic C18 column is 75 μm, the length is 15 cm, and the particle size of C18 is 3 μm. Mobile phase A solution: 0.1% (v) trifluoroacetic acid in water; Mobile phase B solution: 0.1% (v) trifluoroacetic acid in 80% (v) acetonitrile solution; Mobile phase B linear gradient: 5~32% ; Flow rate of mobile phase: 0.3 μL / min.
[0078] (3) The mass spectrometry conditions for proteome analysis were: the fragmentation mode of the mass spectrometer was HCD, the time was 78 min, and the flow rate was 0.3 μL / min. The mass range of the primary mass s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com