Method for identifying purity of Jingke 968 corn hybrids based on SNP markers
A technology for hybrid purity and corn, applied in the field of molecular biology of crops, can solve the problems of heavy workload, easy to be affected by environment or developmental stage, and difficult to high-throughput detection methods.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Determination of SNP molecular markers and primers used to identify the purity of Jingke 968 corn hybrids
[0038] (1) Basic sites: The 384 SNP sites published by "Maize DNA Fingerprint Library and Variety Molecular Identification SNP Core Site Combination-maizeSNP384" (disclosed in Chinese Patent No. 201410756086.0) were used as the starting site set and established SNP-DNA fingerprint data of 335 nationally approved corn hybrids.
[0039] (2) SNP site test and biological feature evaluation: 192 corn hybrids were selected, and DNA was extracted by rapid extraction, CTAB, kits, and multiple methods. Based on the KASP technology system, the DNA verification sites extracted by different methods were used. The typing effect, repeatability, and stability of the points can be used to obtain a set of candidate sites.
[0040] (3) Polymorphism evaluation of SNP locus: using 200 different types of maize inbred lines and 335 maize hybrids data to evaluate the polymorphism pa...
Embodiment 2
[0049] Example 2 Using the SNP molecular marker provided by the present invention to identify the purity of Jingke 968 corn hybrid
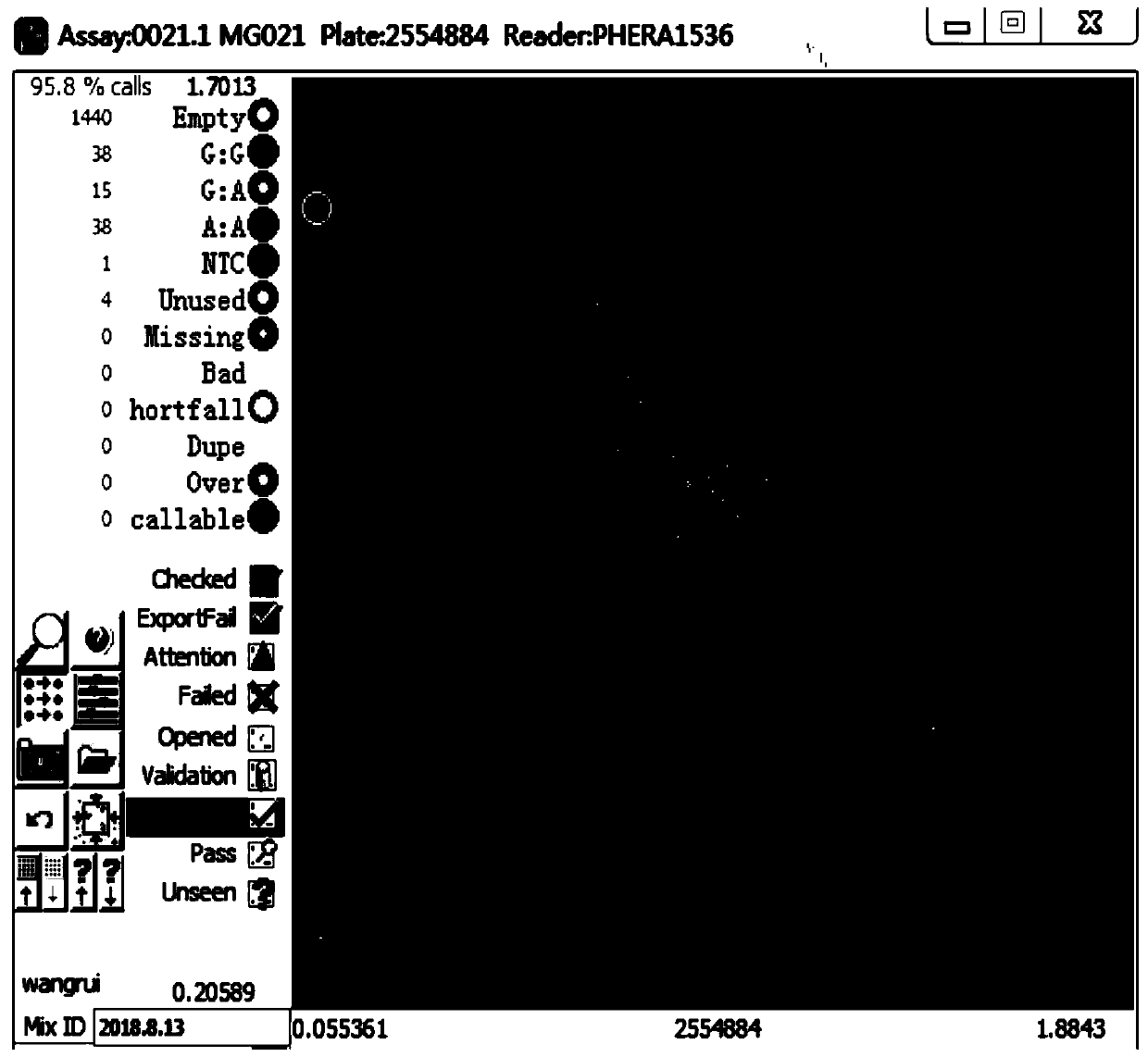
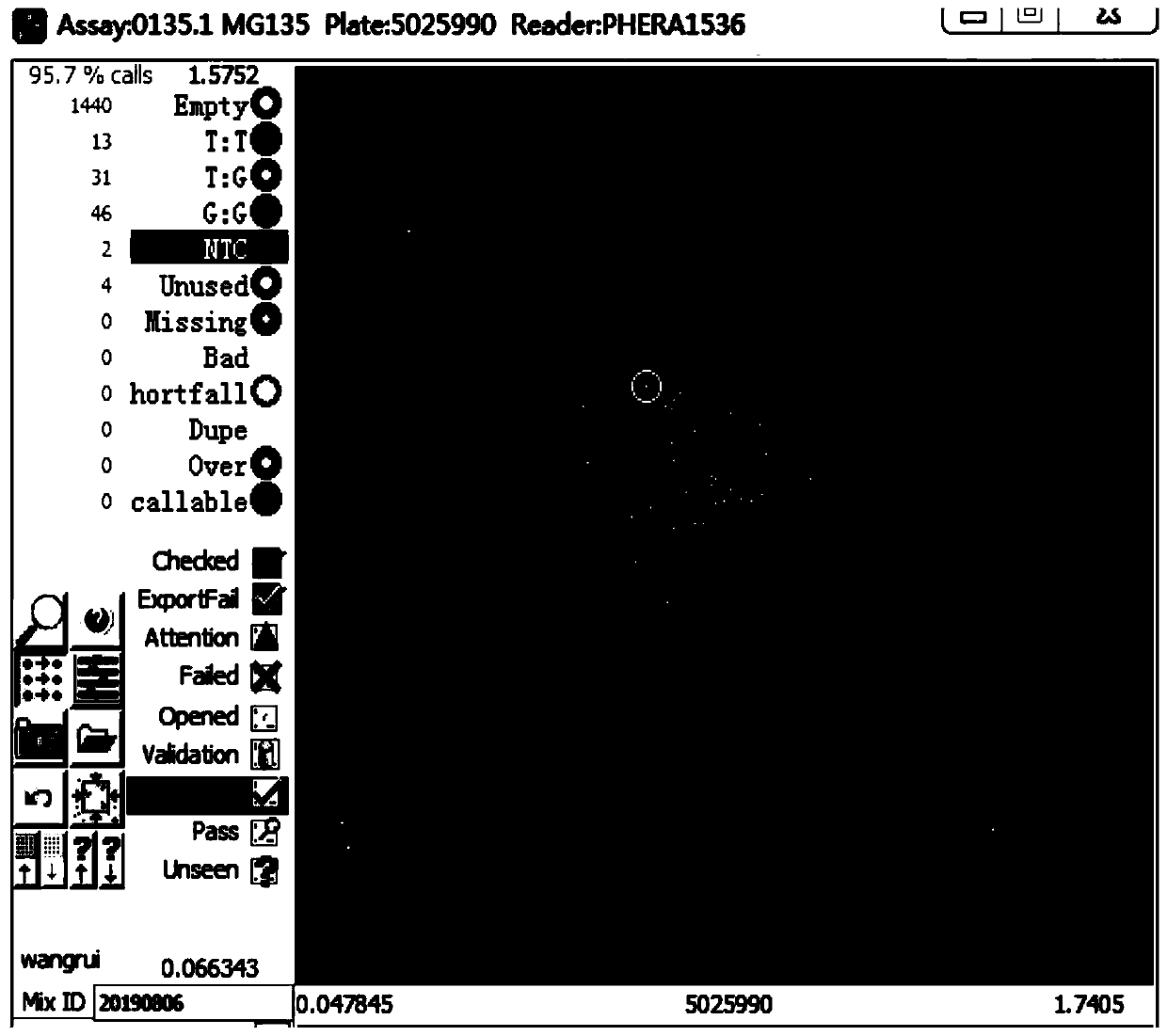
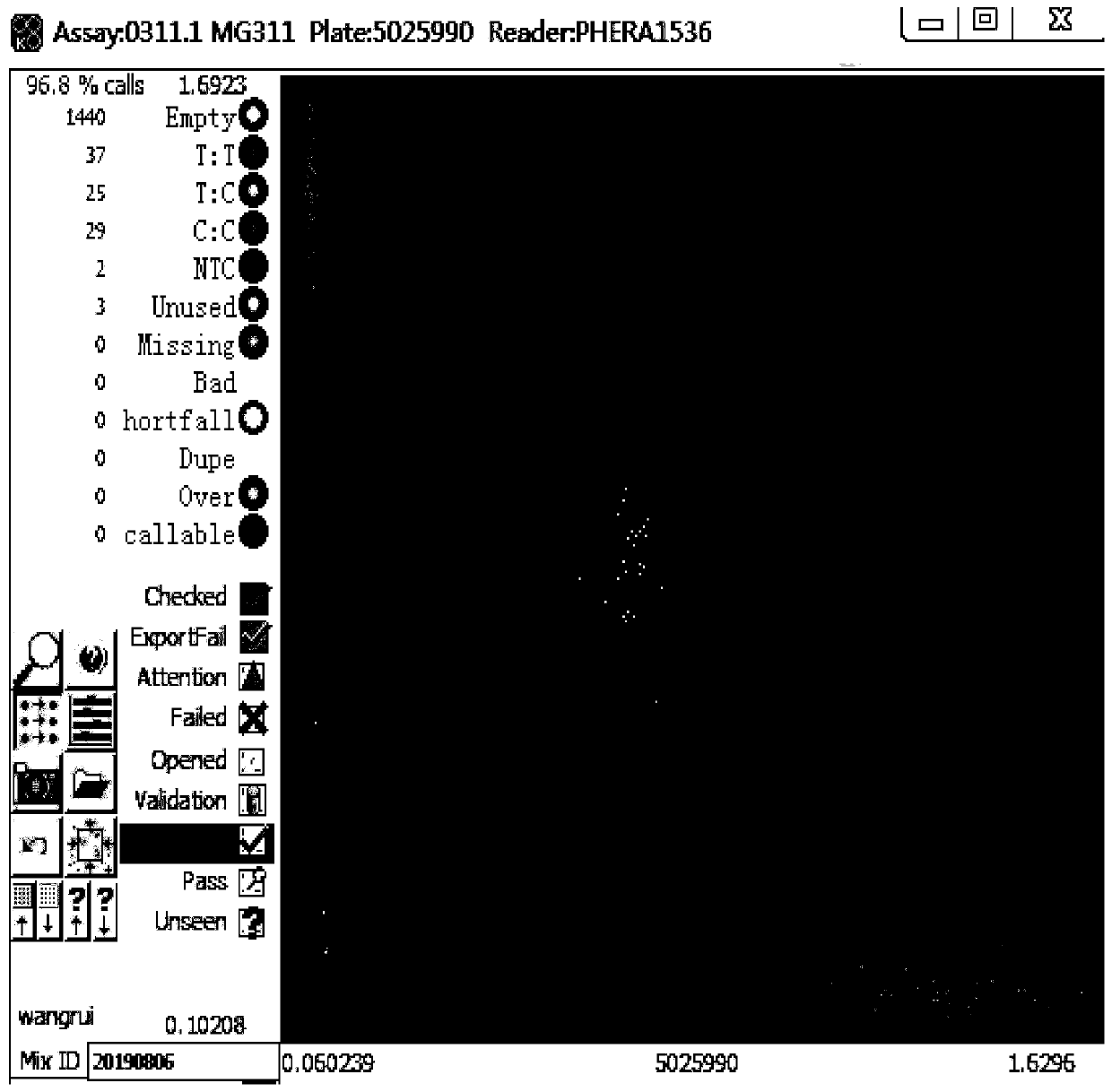
[0050] Using the primer combination designed for 4 SNP sites in Example 1, the scheme for seed purity identification of Jingke 968 is as follows: Synthesize primers according to the KASP technical requirements, and the primers are common primers without fluorescent groups; purchase KASP technology matching PCR amplification system MasterMix; configure the reaction system to add DNA, primers and MasterMix; run the amplification reaction program; scan the fluorescence signal in situ; perform data analysis to obtain genotype data; according to each individual plant of Jingke 968 on each pair of primers Genotype data, determine whether the individual plant is a normal plant, a selfed seedling, or a heterogeneous plant, and finally determine its purity value comprehensively. The specific operations are as follows:
[0051] (1) Preparation of samples to be...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com