Application of OsPTP1 in efficient plant phosphorus breeding
An efficient and versatile technology, applied in the field of plant genetic engineering, can solve the problems of insufficient phosphorus availability, low phosphorus content, and affecting plant growth and development.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1. Research on the interaction of OsPTP1 with OsPT2 and OsPT8:
[0064] Firstly, a protein phosphatase OsPTP1 capable of interacting with OsPT8 was screened by yeast two-hybrid. Its genome sequence is 3892bp (SEQ ID No. 1) and encodes a 290 amino acid protein (SEQ ID No. 2).
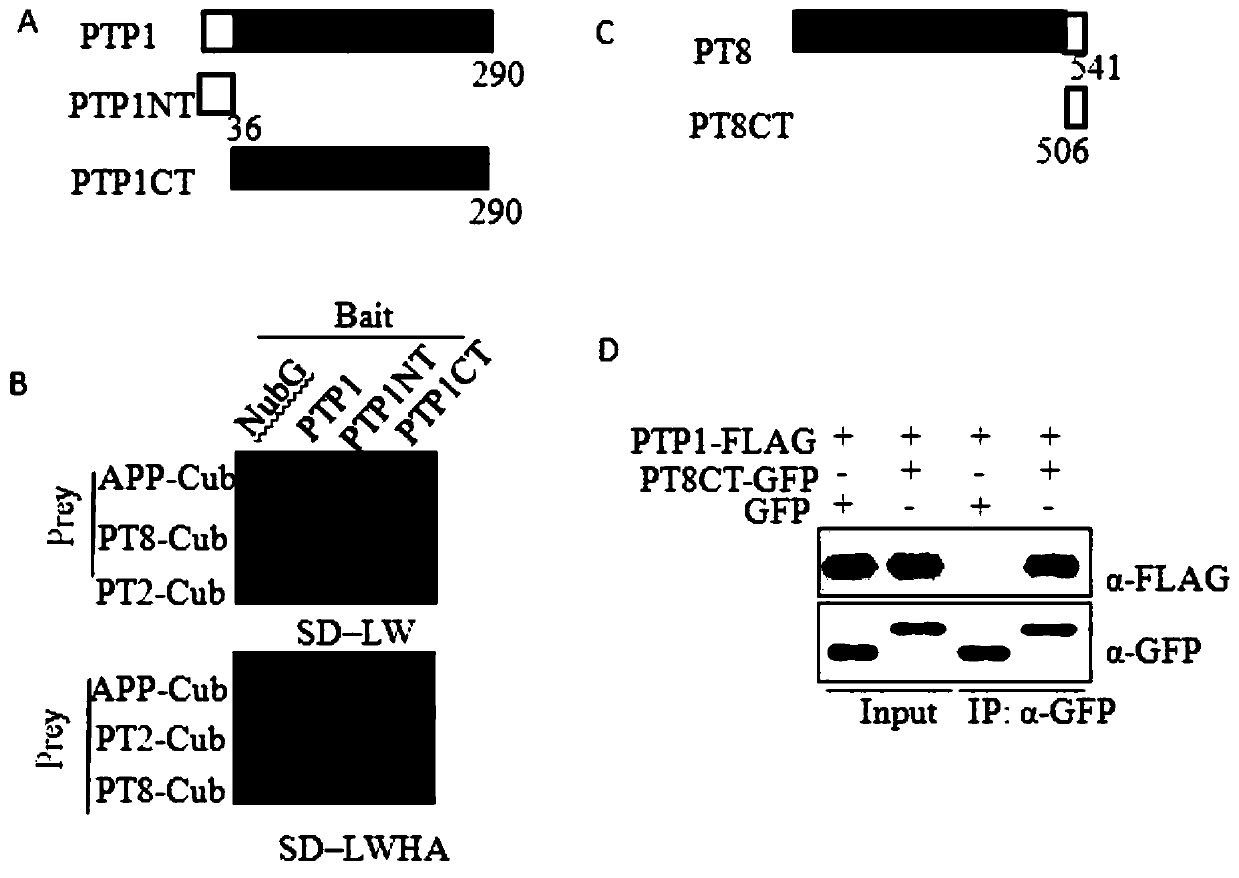
[0065] Then further design confirmed that in addition to OsPT8, OsPT2 can also interact with OsPTP1. By analyzing the protein structure of OsPTP1, it was found that it consists of 36 amino acids at the N-terminus and 254 amino acids at the C-terminal PP2C phosphatase domain ( figure 1 A). In order to clarify the subdomains that interact with OsPT2 and OsPT8, the two subdomains of OsPTP1 were truncated, and it was confirmed by yeast two-hybrid experiments that OsPTP1 interacted with OsPT2 and OsPT8 through its N-terminal 36 amino acids ( figure 1 B). Subsequently, the interaction between OsPTP1 and OsPT8 was further clarified by in vivo Co-IP experiments ( figure 1 C and 1D).
Embodiment 2
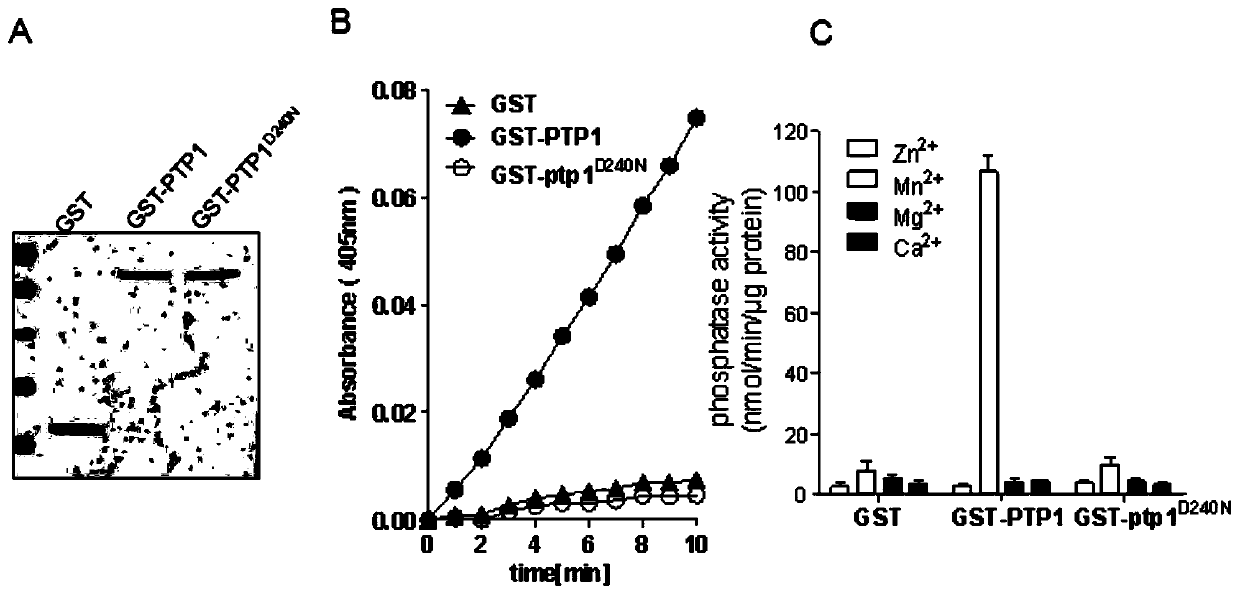
[0066] Example 2. OsPTP1 is verified to have protein phosphatase activity in vitro:
[0067] In order to clarify that OsPTP1 has protein phosphatase activity, it was prokaryotically expressed and purified. Using the wild-type cDNA as a template, the full-length coding sequence of OsPTP1 was amplified with OsPTP1-specific primers, and the target fragment size was 873bp. The amplified target fragment was double-digested with BamH I and Sal I, and ligated into the pGEX4T-1 vector after the same digestion. In order to verify that OsPTP1 has PP2C phosphatase activity, one of the Mg 2+ / Mn 2+ Mutation of the ion binding site (specifically, aspartic acid at the 240th amino acid position of OsPTP1 protein is mutated to asparagine). The pGEX4T-1 vector was constructed by the same ligation method. The primers used are as follows:
[0068] OsPTP1 full-length primer:
[0069]The upstream primer is: CGGGATCCATGGCTGGCAAGGAAATCTACC
[0070] The downstream primer is: GCGTCGACGCAGAGGAAT...
Embodiment 3
[0075] Example 3, construction of OsPTP1-GUS fusion gene vector and GUS staining
[0076] OsPTP1p:OsPTP1:GUS vector construction: use the wild-type Nipponbare genome as a template to amplify the OsPTP1 promoter, 5'UTR and the whole genome sequence (excluding the terminator, 6408bp), use Sal I and BamH I to digest, and ligate into pBI-GUS-PLUS vector after the same digestion; get OsPTP1p:OsPTP1:GUS vector.
[0077] The primer information is as follows:
[0078] Upstream primer: GCGTCGACTAAACGACGGTAAAAATCCCTAAT
[0079] Downstream primer: CGGGATCCTGCAGAGGAATTTTACAACGATGC
[0080] After being verified by sequencing, it was integrated into the wild-type Nipponbare genome through Agrobacterium infection.
[0081] GUS staining and microscopic observation
[0082] Soak the roots, rhizome junctions, stems, leaves, flowers and other organs of T1 generation transgenic seedlings in the GUS dye solution (recipe as shown in Table 1), vacuumize at room temperature for 10 minutes, and th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com