CRISPR/Cas9 vector suitable for Dichotomomycescejpii FS110, construction method and application thereof
A technology of FS110 and construction method, applied in the fields of biochemistry and molecular biology, can solve problems that have not yet been seen, and achieve the effect of promoting development and utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Construction of targeted gliotoxin biosynthesis gene knockout vector
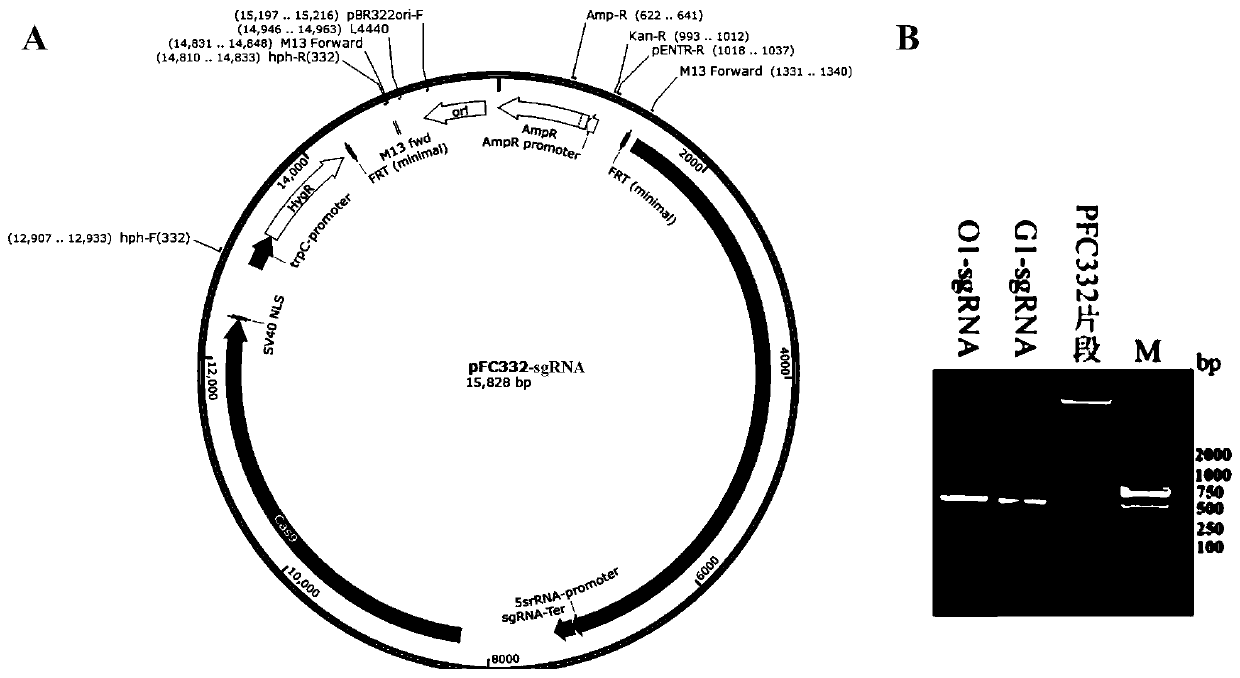
[0033] On the basis of the pFC332 plasmid, pFC332 was subjected to double enzyme digestion at the PacI and BglII sites, and a sequence was inserted: 5SrRNA promoter-targeting sequence-sgRNA-sgRNA terminator (including the 5SrRNA promoter applicable in Eideria FS110 , the sgRNA targeting sequence backbone gene and its terminator that function in yeast cells), so that the plasmid can stably express Cas9 protein and transcribe sgRNA at the same time after being transformed into protoplasts (the plasmid is named pFC332-sgRNA).
[0034] The primers designed for constructing the vector are shown in Table 1, and the pFC332-sgRNA vector was constructed by restriction enzyme ligation. The construction method is as follows:
[0035] Table 1 Primer Sequence
[0036]
[0037]Using Eydleria FS110 genomic DNA as a template, and using primers 5SrRNA-promoter-F and 5SrRNA-promoter-R as primers before...
Embodiment 2
[0043] Knockout of gliotoxin biosynthesis genes in E. dermatella FS110:
[0044] The method of introducing exogenous gene into the protoplast of Eideria FS110 is as follows:
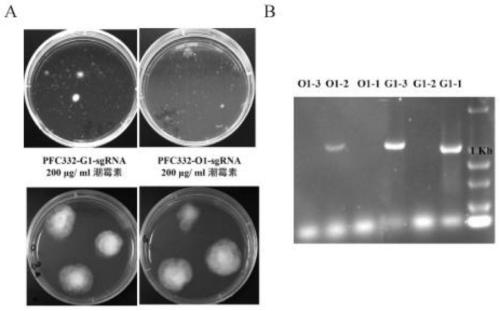
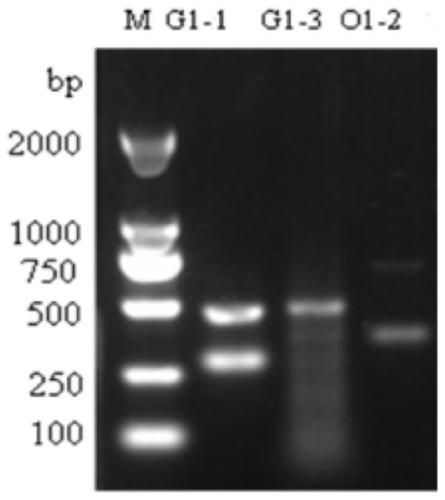
[0045] (1) Protoplasts of Eedleia FS110 prepared (for the specific preparation method refer to the inventor's patent number 201510540618.1, the name is: a patent of Eedleia FS110 protoplasts and its preparation method and transformation method) (1×10 8 / mL) were mixed with 2.5-5 μg G1-pFC332-sgRNA plasmid or O1-pFC332-sgRNA plasmid, and placed on ice for 5 minutes, then added 200 μL volume fraction of 30% PEG4000, placed at 30 ° C for 15 minutes, and then added 400 μL PEG4000, Place at 30°C for 15 minutes, then add 1.2mL of W5 solution to terminate the reaction, and finally add 4mL of WI buffer and place it on a shaker at 30°C at low speed for overnight incubation;
[0046] (2) After cooling the melted TB3 solid medium to room temperature, take 20 mL each time and mix gently with the overnight culture s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com