Primer group for rapid detection of 2019-nCoV based on CRISPR technology and application thereof
A 2019-ncov, detection primer technology, used in microorganism-based methods, recombinant DNA technology, and microbial assay/inspection. The effect of advantage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Design of primer sequences for 2019-nCoVCRISPR detection.
[0057] 1. Target sequence selection.
[0058] Novel coronavirus (2019-nCoV) is a newly discovered coronavirus. The inventor analyzed the sequence of 2019-nCoV and believed that Orf1ab gene and N gene are conserved genes of coronavirus, which can be designed according to the conserved sequence, according to the target Spot the Orf1ab gene and N gene sequence regions, design crRNA and RPA amplification primers, and test the sensitivity of the detection method.
[0059] The sample to be tested in this embodiment is a plasmid (synthesized by Aiji Biotechnology Co., Ltd.) inserted with a selected target sequence region of 2019-nCoV. The sequence of Orf1ab gene insertion is as follows:
[0060] 5’-ATGCCTTCAAACTCAACATTAAATTGTTGGGTGTTGGTGGCAAACCTTGTATCAAAGT AGCCACTGTACAGTCTAAAATGTCAGATGTAAAGTGCACATCAGTAGTCTTACTCTCAGT TTTGCAACAACTCAGAGTAGAATCATCATCTAAATTGTGGGCTCAATGTGTCCAGTTACA CAATGACATTCTCTTAGCTAAAGATACTACTGAAGCCTTT...
Embodiment 2
[0072] 1. The amplification efficiency screening of RPA amplification primers.
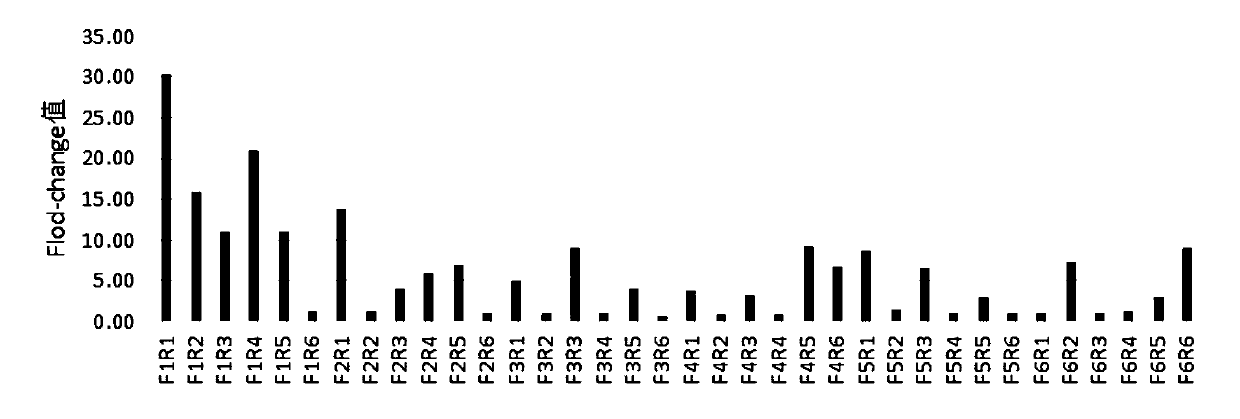
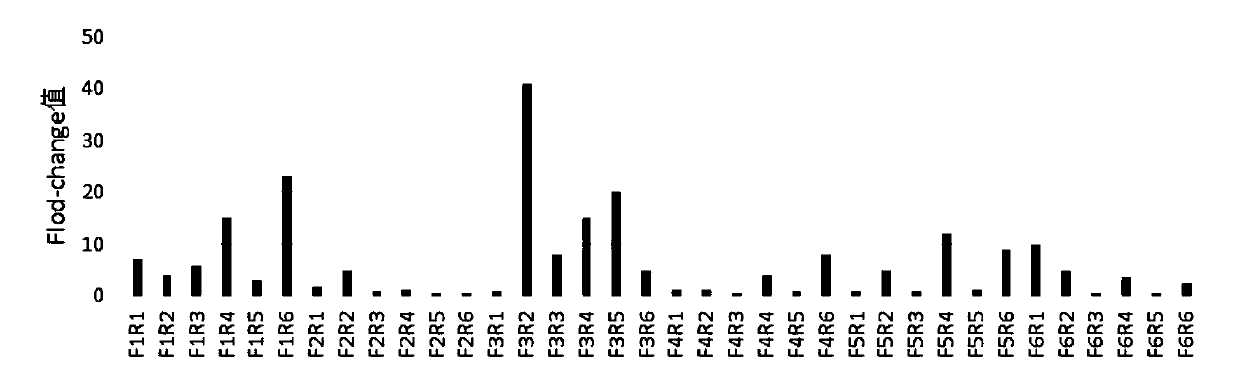
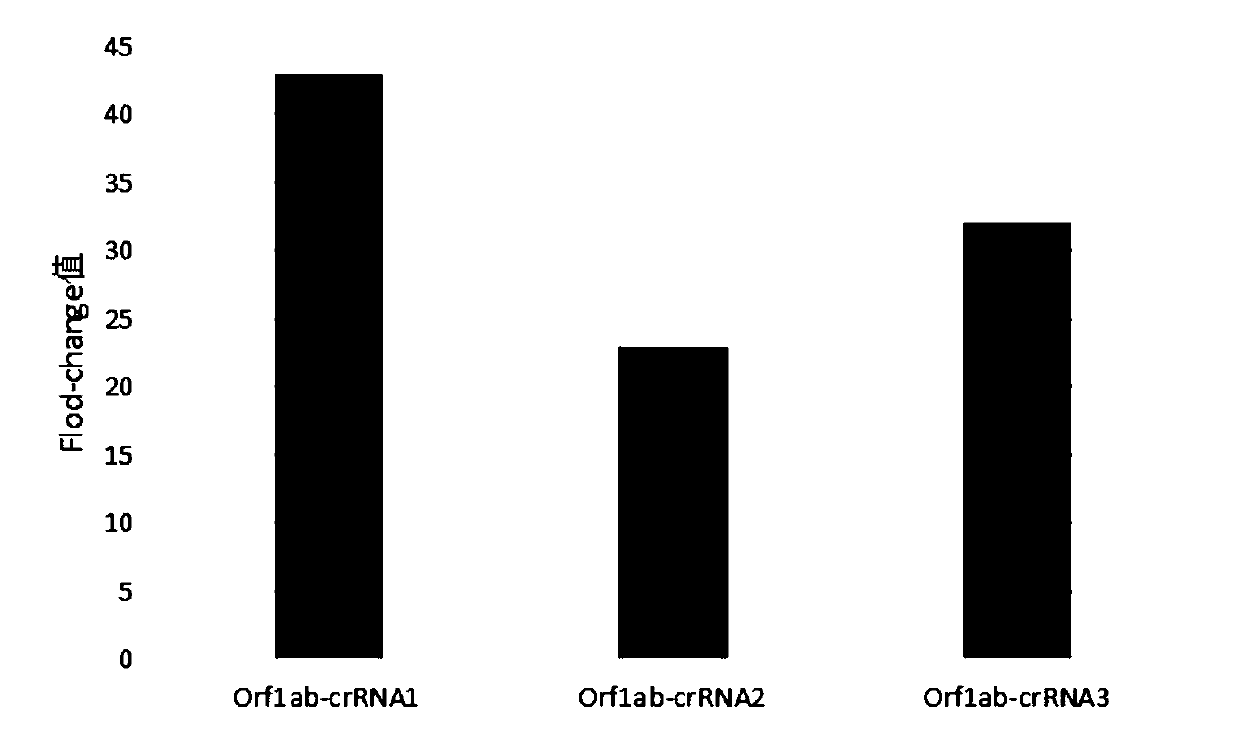
[0073]In order to screen the RPA amplification primers of Cas13a, the plasmids inserted with the gene sequences of the two 2019-nCoV targets Orf1ab gene and N gene in the above-mentioned Example 1 were used as standard samples, and extracted according to conventional methods to obtain standard samples Genes, primers for the target Orf1ab gene (primers shown in the above table 1) are combined in two groups with Orf1ab-crRNA-1, and primers for the target N gene (primers shown in the above table 2) are paired Combined, and combined with N-crRNA-1 for detection and screening, the template concentration was 1000copies / μl.
[0074] 1.1 Method.
[0075] Reaction system: upstream primer (0.1-0.6μM) 0.5-2.5μL, downstream primer (0.1-0.6μM) 0.5-2.5μL, RPA enzyme master mix 21μL, magnesium acetate (10-20mM) 0.5-2μL, signal reporter probe (50-400nM) 1μl, NTP mixed solution (0.2-6mM) 1-10μL, T7 RNA polymeras...
Embodiment 3
[0097] In this example, sensitivity detection is performed based on RPA amplification, T7 in vitro transcription and Cas13a.
[0098] Using the plasmids with the conserved sequence of the nCoV-Orf1ab gene and the conserved sequence of the nCoV-N gene as templates, the calculated dilutions are 3000copies / μL, 300copies / μL, 30copies / μL, 3copies / μL, 1copy / μL, 0.4copy / μL , 0.16copy / μL, a total of 7 gradients were used as templates for sensitivity detection, and the amount of template added to each reaction was 2.5 μL.
[0099] 1. Method.
[0100] Referring to the method in Example 2 above, the sensitivity analysis was performed on the nCoV-Orf1ab target and the nCoV-N target respectively, and a negative control was set in the experiment.
[0101] Reaction conditions: react at 42°C for 40-60 minutes, and read the fluorescence value of FAM every 1 minute.
[0102] Refer to the result interpretation method in the above-mentioned embodiment 2 to perform result interpretation.
[010...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Upstream primer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com