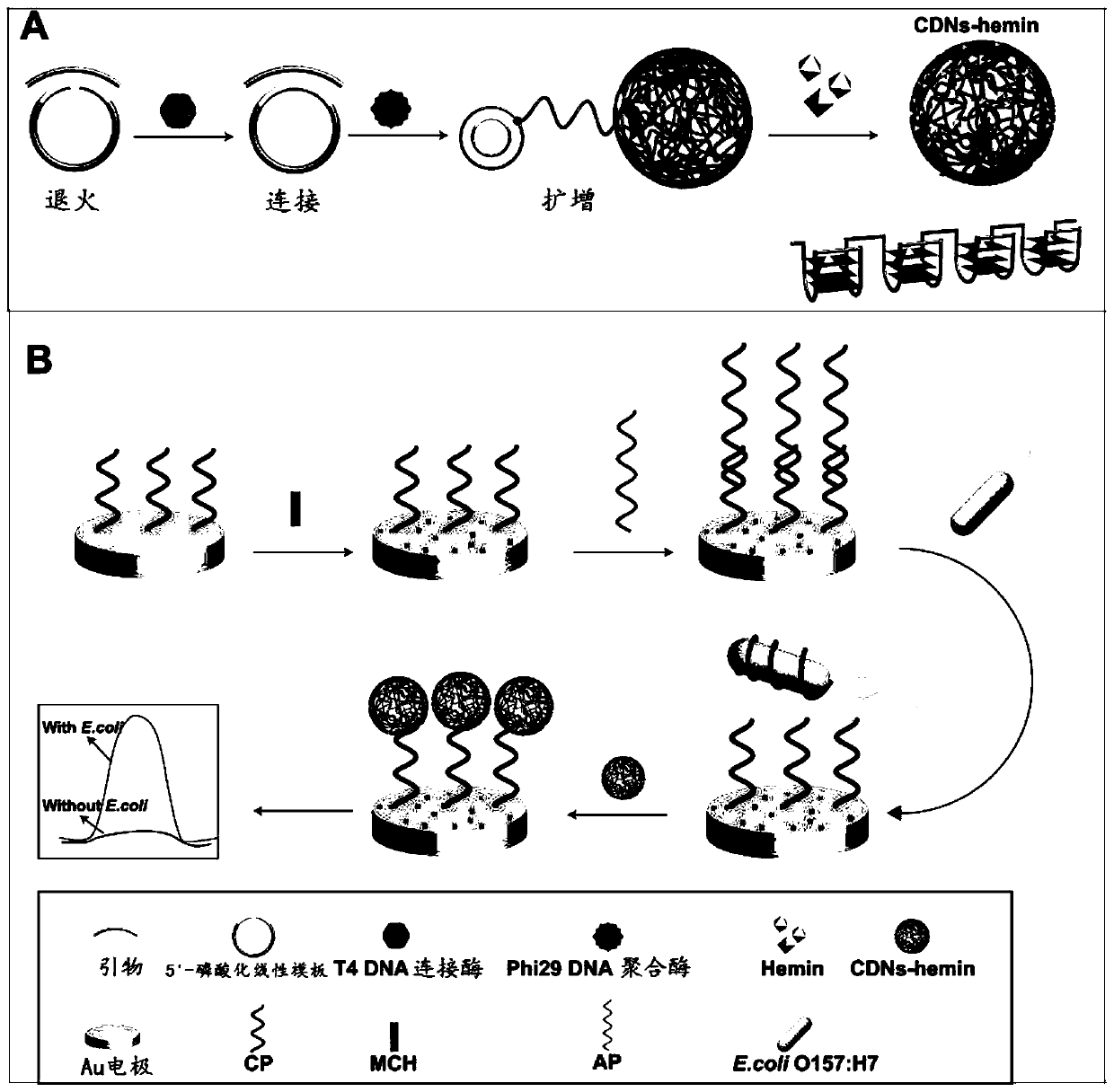
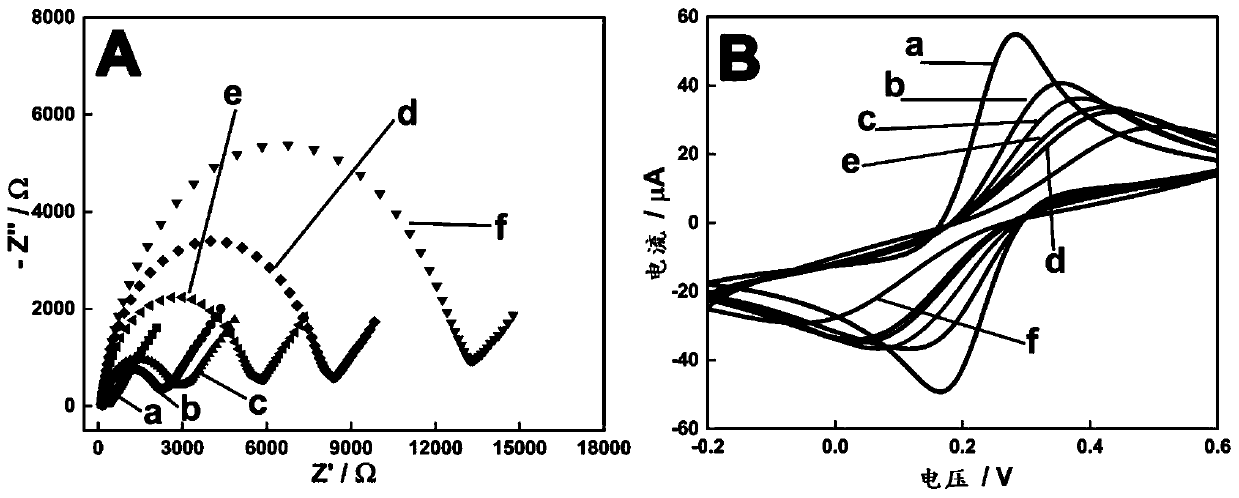
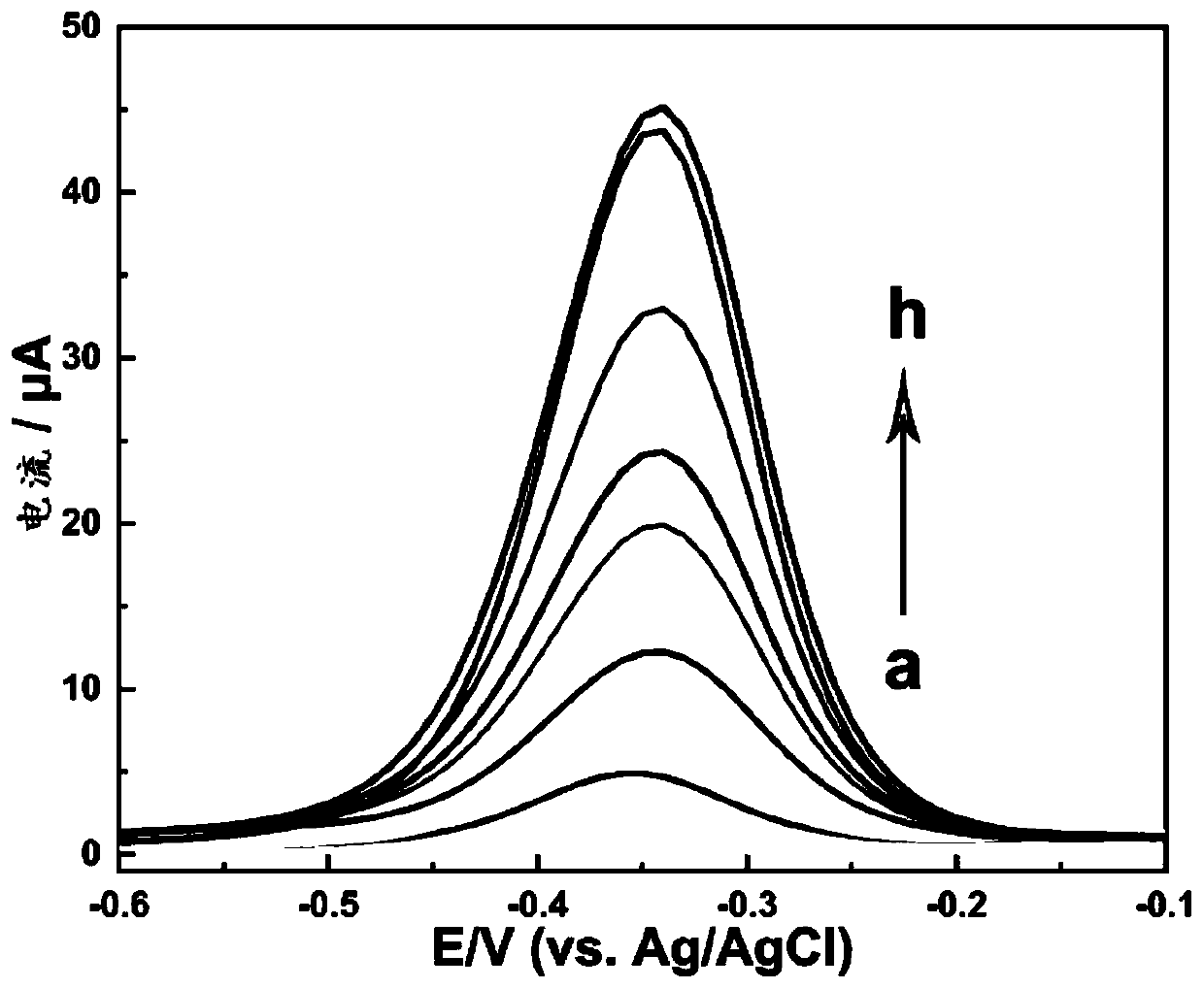
DNA nanostructure, electrochemical aptamer biosensor system and preparation method and application of electrochemical aptamer biosensor system
A nanostructure and aptamer technology, applied in the direction of material electrochemical variables, scientific instruments, instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0066] A kind of preparation method of DNA nanostructure of the present invention, mainly is to synthesize DNA nanostructure (RCA NCs) by rolling circle amplification reaction, comprises the following steps:
[0067] Step 1. Annealing
[0068] In T4 DNA ligase buffer (40mM Tris-HCl, 10mM MgCl 2 , 10mM DTT, 0.5mM ATP, pH7.8), mix the 5'-phosphorylated linear template with the primer, heat the mixture and then cool it to room temperature to form a circular template-primer hybrid;
[0069] Wherein, the nucleic acid sequence of the 5'-phosphorylated linear template is 5'-(PO 4 3- )-ACCCGCCCTACCCAAAATATGCCCTCGGTTGTGGTATTATACGGCATTCCTCTCCGGACAACCCTCGCGGCATCTCGTCCACACTGCCTAAATTTTCCCA-3';
[0070] Wherein, the nucleic acid sequence of the primer is 5'-TTTTGGGTAGGGCGGGTTGGGAAAA-3';
[0071] Step 2. Connect
[0072] Mix the circular template-primer hybrid with T4 DNA ligase and incubate to ligate the circular template gap;
[0073] Step 3. Enzyme inactivation
[0074] After heati...
Embodiment 1
[0111] Synthesis and preparation of embodiment 1 DNA nanostructure
[0112] (1) Annealing
[0113] In T4 DNA ligase buffer (40mM Tris-HCl, 10mM MgCl 2 , 10mM DTT, 0.5mM ATP, pH7.8), mixed 0.3μM 5'-phosphorylated linear template with 0.6μM primer, heated the mixture at 90°C for 5min, then gradually cooled to room temperature within 3h, to form a circular template-primer hybrid;
[0114] The nucleic acid sequence of the 5'-phosphorylated linear template is 5'-(PO 4 3- )-ACCCGCCCTACCCAAAATATGCCCTCGGTTGTGGTATTATACGGCATTCCTCTCCGGACAACCCTCGCGGCATCTCGTCCACACTGCCTAAATTTTCCCA-3';
[0115] The nucleic acid sequence of the primer is 5'-TTTTGGGTAGGGCGGGTTGGGAAAA-3'.
[0116] (2) connection
[0117] Mix the circular template-primer hybrid with 10 U of T4 DNA ligase and incubate at 16°C for 16 hours to ligate the circular template gap.
[0118] (3) Enzyme inactivation
[0119] Heat at 65°C for 10 min to inactivate T4 DNA ligase.
[0120] (4) Amplification
[0121] Add the 2mM clos...
Embodiment 2
[0126] Hemin / G-quadruplex DNAzyme functionalization of Example 2RCA NCs
[0127] (1) Treat 25 μL of TE buffer containing RCA NCs at 90°C for 1 min, cool to 4°C, and keep for 60 min.
[0128] (2) Add the product obtained in step (1) together with 100 μM Hemin (heme) solution to 50 μL Tris-HCl buffer, and incubate at 37°C for 60 min, so that Hemin (heme) can be embedded in the RCA NCs In the G-quadruplex unit, functionalized RCA NCs were formed, that is, functionalized Hemin / G-quadruplex DNAzyme.
[0129] (3) Store the prepared functionalized Hemin / G-quadruplex DNAzyme below 4°C. RCA NCs with peroxidase-mimicking DNAzyme activity can catalyze the substrate H 2 o 2 reduction, resulting in an electrochemical signal.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com