PCR-HRM (Polymerase Chain Reaction-High Resolution Melting) primer for authenticating swine acute diarrhoea syndrome virus and porcine epidemic diarrhea virus, method for authenticating swine acute diarrhoea syndrome and porcine epidemic diarrhea, and application of PCR-HRM primer
A PCR-HRM, porcine epidemic diarrhea technology, applied in microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve problems such as timely diagnosis and treatment of unfavorable diseases, cumbersome operation methods, economic losses, etc., and achieve rapid differentiation. , good repeatability, shorten the time effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1PCR-HRM primer design
[0042] A primer pair P-F and P-R for amplifying partial gene sequences of porcine acute diarrhea syndrome virus and porcine epidemic diarrhea virus were designed according to the conserved gene sequences of porcine acute diarrhea syndrome virus and porcine epidemic diarrhea virus, and their base sequences are as follows:
[0043] P-F: GATGGTGGTTGTAASACTAT;
[0044] P-R: ACAGTRGCACCTATGTAGCC.
[0045] One degenerate base S (equivalent to G / C) was introduced into primer P-F, and one degenerate base R (equivalent to A / G) was introduced into P-R.
Embodiment 2
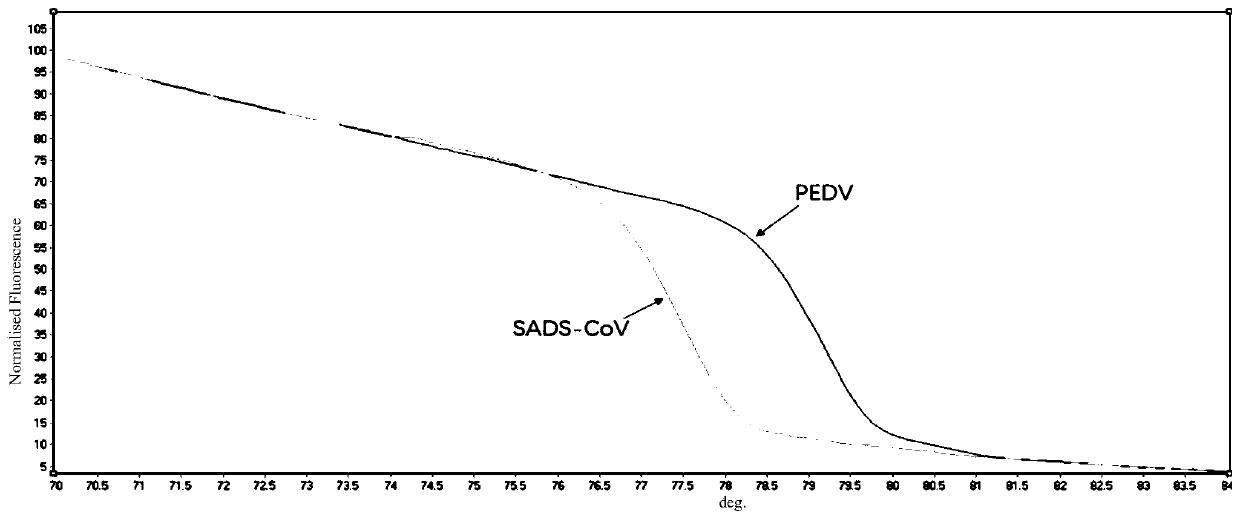
[0046] The preparation of embodiment 2 standard sample and PCR-HRM analysis thereof
[0047] (1) Extraction of viral RNA: use AxyPrep TM The bodily fluid virus DNA / RNA preparation kit (product serial number: PID0320425) was used to extract the RNA of porcine acute diarrhea syndrome virus and porcine epidemic diarrhea virus in diseased material samples (collected from a pig farm in Guangdong Province in 2019). Disease material samples are mainly intestinal and other tissue samples.
[0048] (2) Reverse transcription to obtain cDNA: use the RNA obtained in the previous step as a template, add reverse transcription reagents, and put the PCR tube into the PCR instrument for the reaction system shown in Table 1, and reverse transcribe at 37°C for 15 minutes; Inactivate at 85°C for 5s, and store at -20°C for later use.
[0049] Table 1 reverse transcription PCR reaction system
[0050]
[0051] For the reverse transcription system, first add the first three reagents to the PCR t...
Embodiment 3
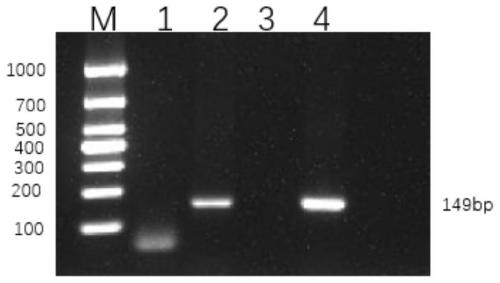
[0068] Embodiment 3 specificity experiment
[0069] Extract the RNA of viruses that commonly cause diarrhea in other pigs, such as porcine transmissible gastroenteritis virus (Porcine Transmissible Gastroenteritis Virus, TGEV) (TGEV-SY strain, which has been published in the literature "You Liuyang et al., Porcine Transmissible Gastroenteritis Virus SY strain Separation and identification of S gene and sequence analysis of S gene. China Veterinary Science, 2016.46(05): pp. 579-585." published in), Porcine Deltacoronavirus (Porcine Deltacoronavirus, PDCoV) (H223 strain, has been published in the literature "He Dongsheng et al., Isolation and identification of porcine delta virus H223 strain and continuous passage and genetic evolution analysis of N gene. Porcine Science, 2019.36(07): pp. 72-74.") and porcine rotavirus (Porcine Rotavirus, PoRV ) (porcine transmissible gastroenteritis, porcine epidemic diarrhea, porcine rotavirus (G5 type) triple live vaccine (attenuated Sinoviru...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com