Kit for detecting novel-coronavirus-susceptive gene and platform
A technology of susceptibility genes and kits, which is used in the determination/examination of microorganisms, resistance to vector-borne diseases, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: The primers for detecting virus susceptibility gene mutations were synthesized by Shanghai Bailige Biotechnology Co., Ltd., and the sequences are as follows:
[0042] Amplification primers:
[0043]
[0044]
[0045] Single base extension primers:
[0046]
[0047]
Embodiment 2
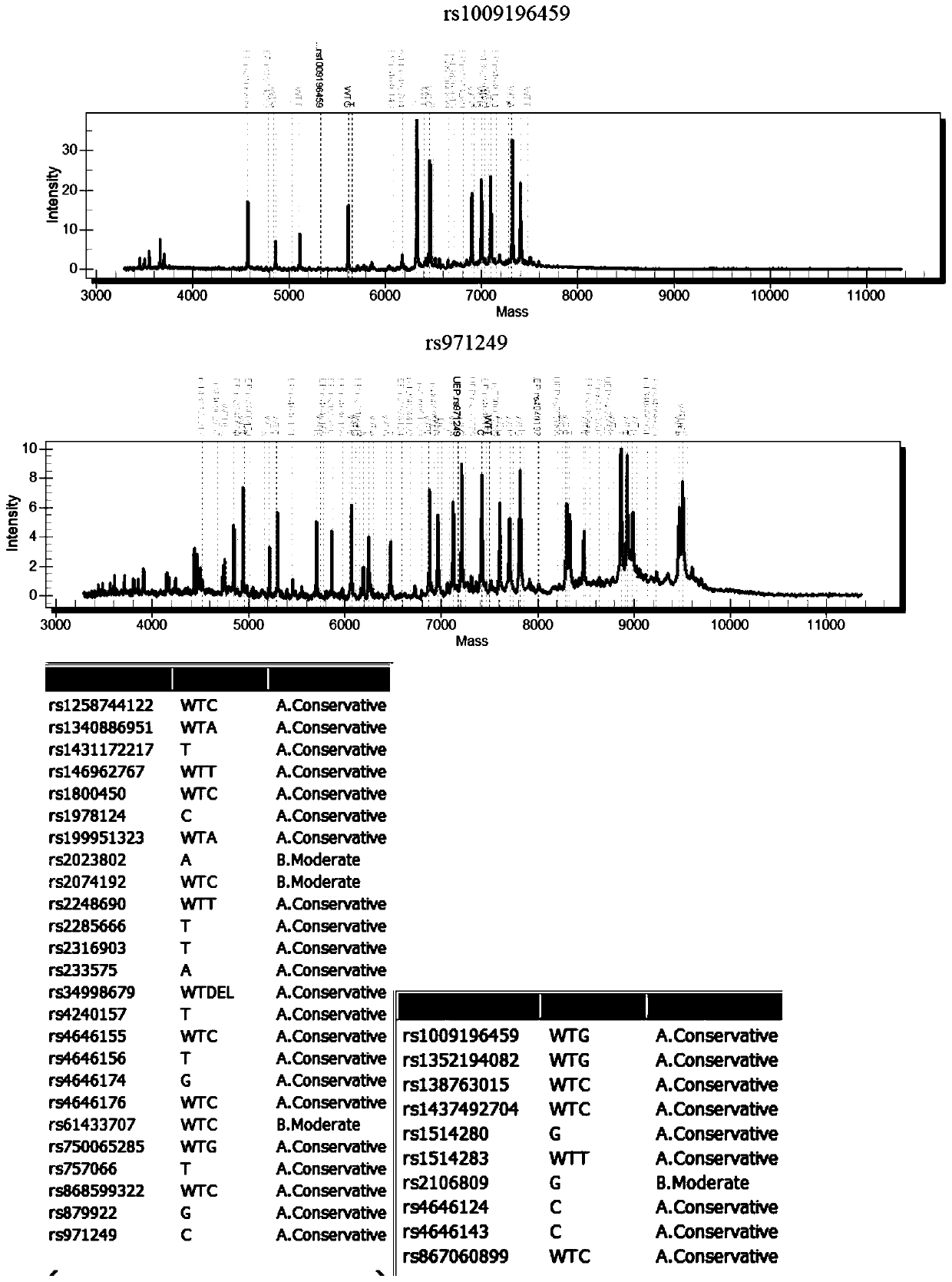
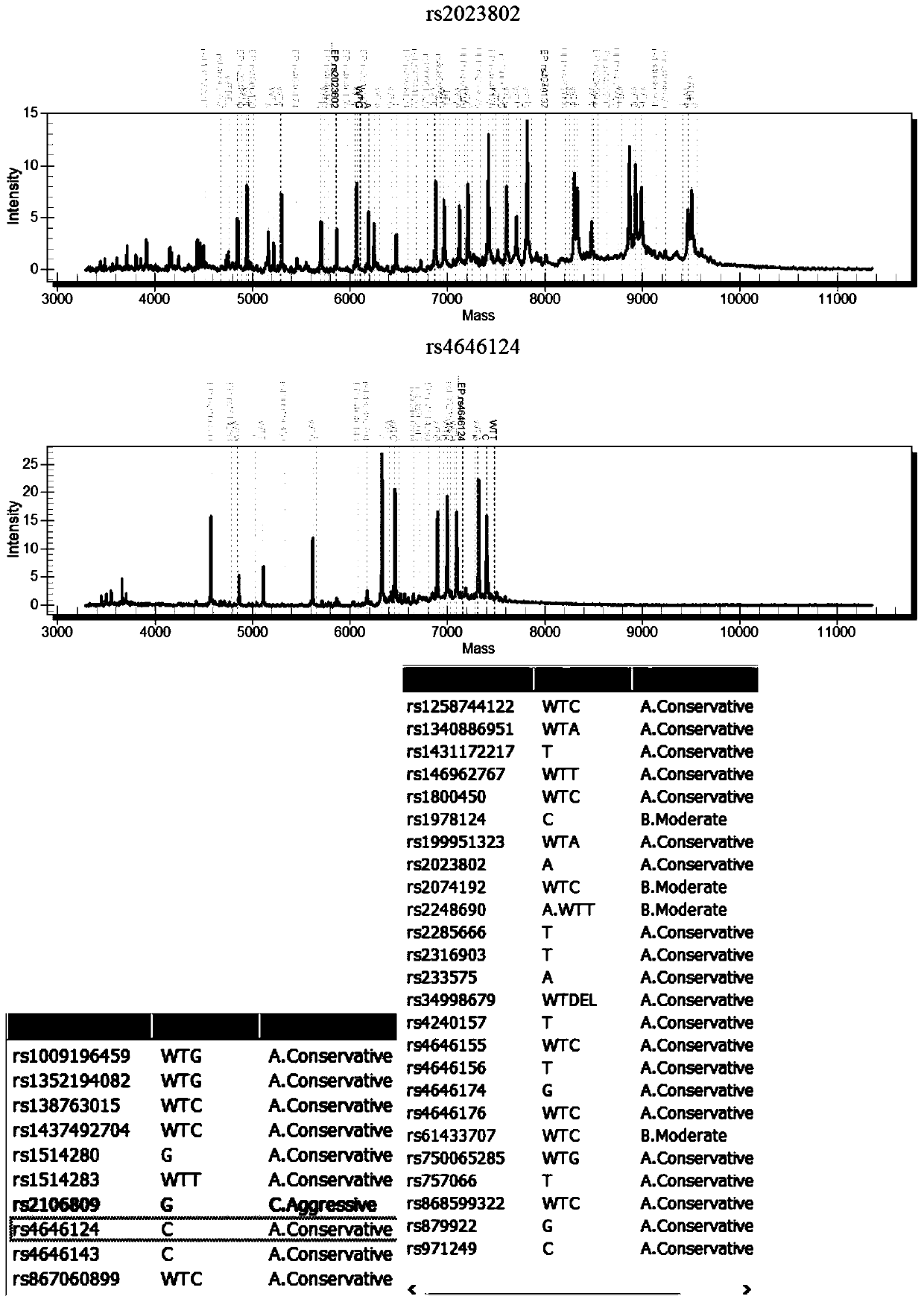
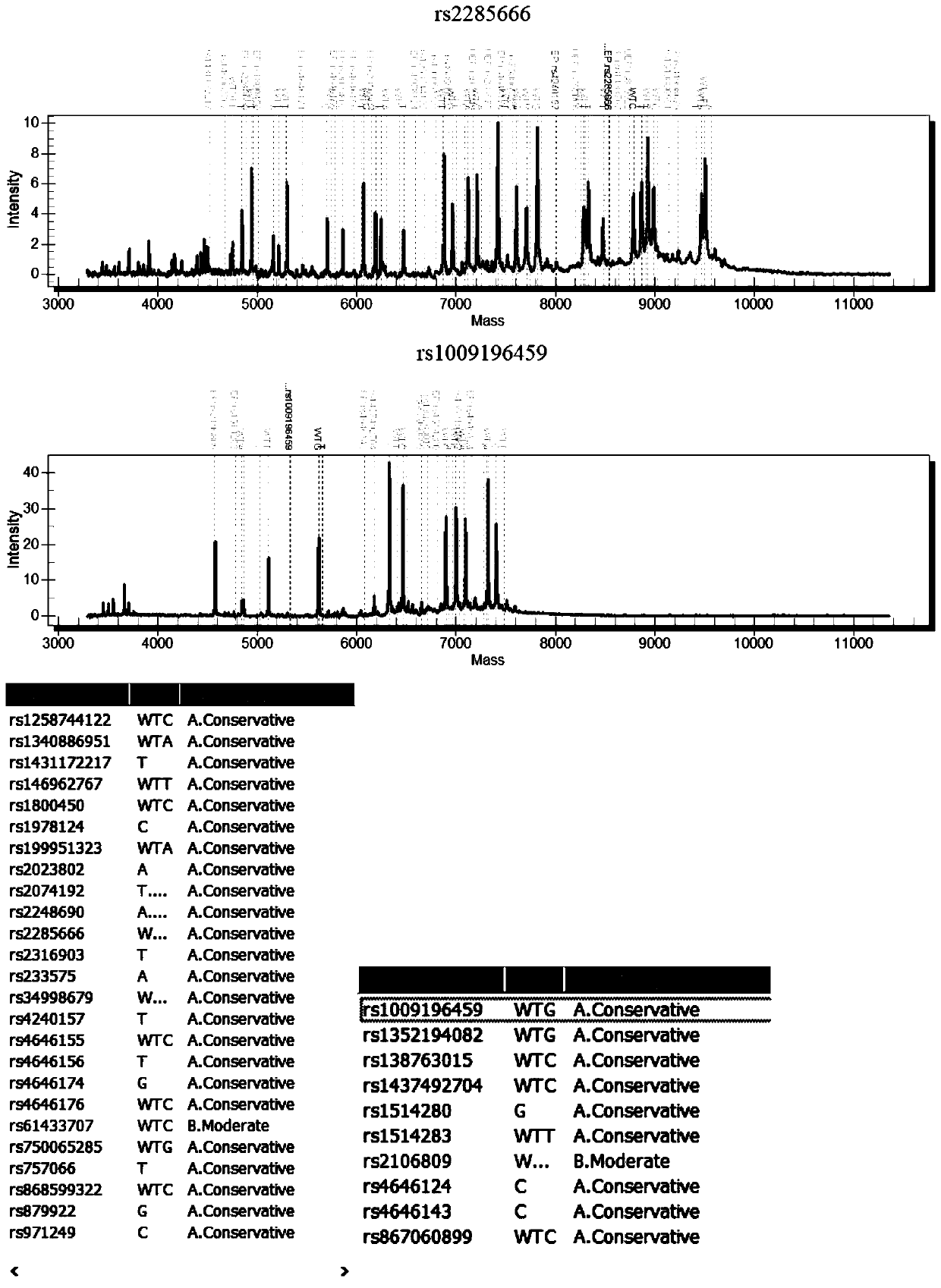
[0048] Example 2: The mutant plasmids for detecting virus susceptibility genes were synthesized by Shanghai Sangon Bioengineering Co., Ltd., and there were 5 mutant plasmids (pUC57 clone plasmid+insert sequence), as shown in the following table:
[0049]
[0050]
[0051] Note: *The SNP number, common name and mutation frequency data of the corresponding mutation sites come from the global shared SNP database NCBI dbSNP (https: / / www.ncbi.nlm.nih.gov / snp / ), OMIM database (http: / / www.omim.org / ), the International 1000 Genomes SNP Database (https: / / www.ncbi.nlm.nih.gov / variation / tools / 1000genomes / ), literature, and relevant domestic guidelines and reports in China.
Embodiment 3
[0052] Example 3: Preparation of a mutation detection kit for virus susceptibility genes.
[0053] (1) Time-of-flight mass spectrometry detection system nucleic acid sample pretreatment reagent (Zhejiang Dipu Diagnostic Technology Co., Ltd., article number: 20010100), including the following main components:
[0054]
[0055] (2) Amplification reaction primer premix: a mixture of the nucleotide sequences shown in SEQ ID NO: 1 to 38 in claim 1; the preferred solution is that the primers shown in SEQ ID NO: 1 to 11 The concentration is 0.5 μM, the concentration of primers shown in SEQ ID NO: 12-25 is 1 μM, and the concentration of primers shown in SEQ ID NO: 26-38 is 2 μM;
[0056] (3) Single-base extension reaction primer premix: a mixture of nucleotide sequences shown in SEQ ID NO: 39 to 73 in claim 2, the preferred molar concentration of each extension primer is as follows:
[0057] SEQ ID No.39-42: SEQ ID No.43-46: SEQ ID No.47-49: SEQ ID No.50: SEQ ID No.51-52: SEQ ID N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com