Methods and compositions for improving engineered microbes
A composition and genetic engineering technology, applied in biochemical equipment and methods, botanical equipment and methods, fertilization methods, etc., can solve problems such as unclear pathways
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
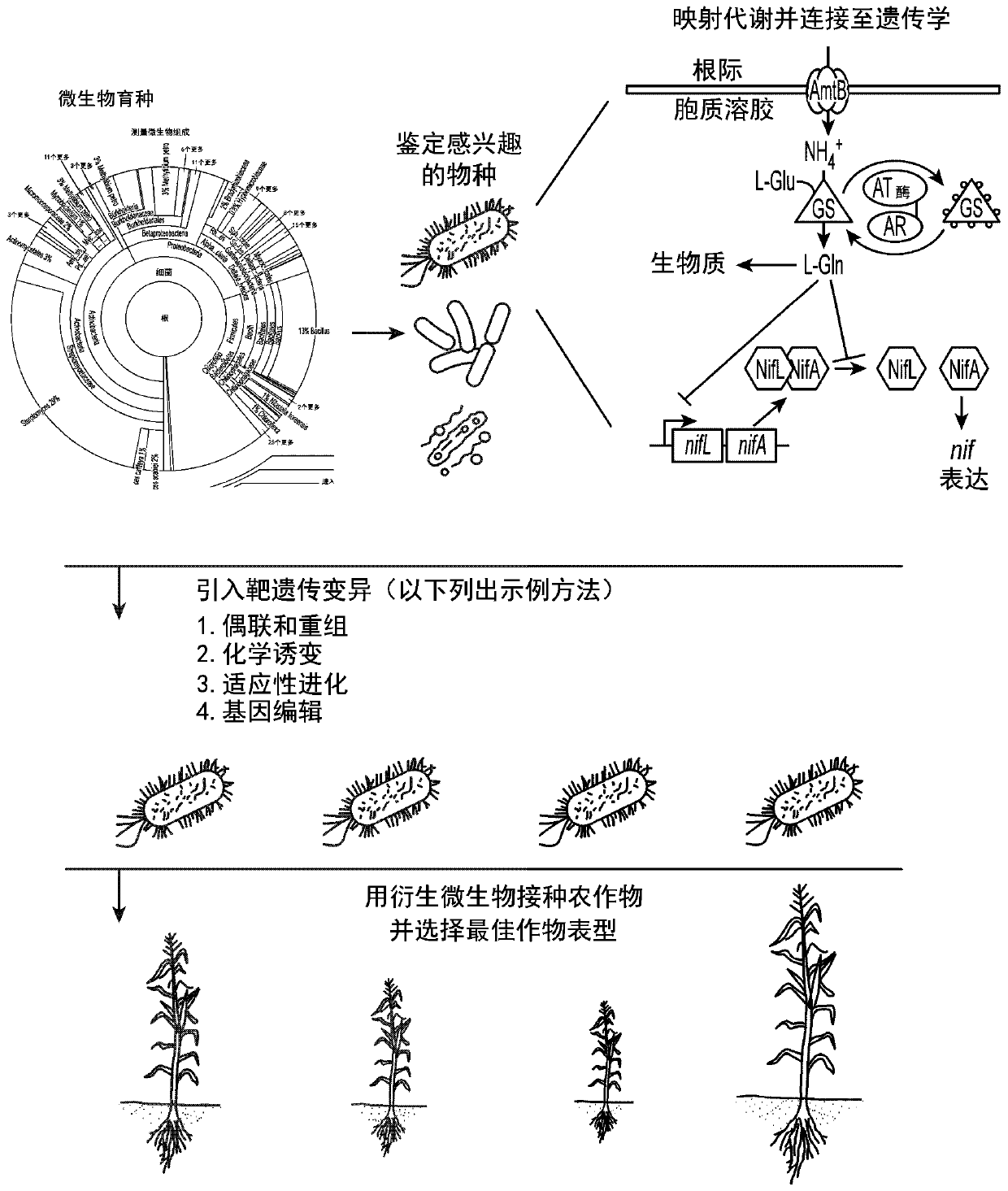
[0315] Example 1: Transcriptome profiling of candidate microorganisms
[0316] Transcriptome profiling of strain CI010 was performed to identify promoters active in the presence of ambient nitrogen. Strain CI010 was grown in defined nitrogen-free medium supplemented with 10 mM glutamine. Total RNA was extracted from these cultures (QIAGEN RNeasy kit) and subjected to RNAseq sequencing by Illumina HiSeq (SeqMatic, Fairmont, CA). Geneious was used to map sequencing reads to CI010 genomic data and identify highly expressed genes under the control of proximal transcriptional promoters.
[0317] Tables 3 and 4 list the genes and their relative expression levels measured by RNASeq sequencing of total RNA. Sequences of proximal promoters are recorded for mutagenesis of nif pathways, nitrogen utilization-related pathways, colonization-related pathways, or genes with desired expression levels.
[0318] table 3
[0319]
[0320]
[0321] Table 4
[0322]
Embodiment 2
[0323] Example 2: Mutagenesis of Candidate Microorganisms
[0324] λ-Red-mediated knockout
[0325] Several mutants of candidate microorganisms were generated using plasmid pKD46 or derivatives containing a kanamycin resistance marker (Datsenko et al. 2000; PNAS 97(12):6640-6645). A knockout cassette was designed with 250 bp of homology flanking the target gene and generated via overlap extension PCR. Candidate microorganisms were transformed with pKD46, grown in the presence of arabinose to induce λ-Red mechanical expression, ready for electroporation, and transformed with the knockout cassette to generate candidate mutagenized strains. As shown in Table 5, thirteen candidate mutants of the nitrogen fixation regulatory genes nifL, glnB and amtB were generated using four candidate microorganisms and one laboratory strain, Klebsiella oxytoca M5A1.
[0326]
[0327] Table 5: List of individual knockout mutants generated by λ-Red mutagenesis
[0328] Oligonucleotide-directe...
Embodiment 3
[0332] Example 3: In-Plant Phenotypes of Candidate Microorganisms
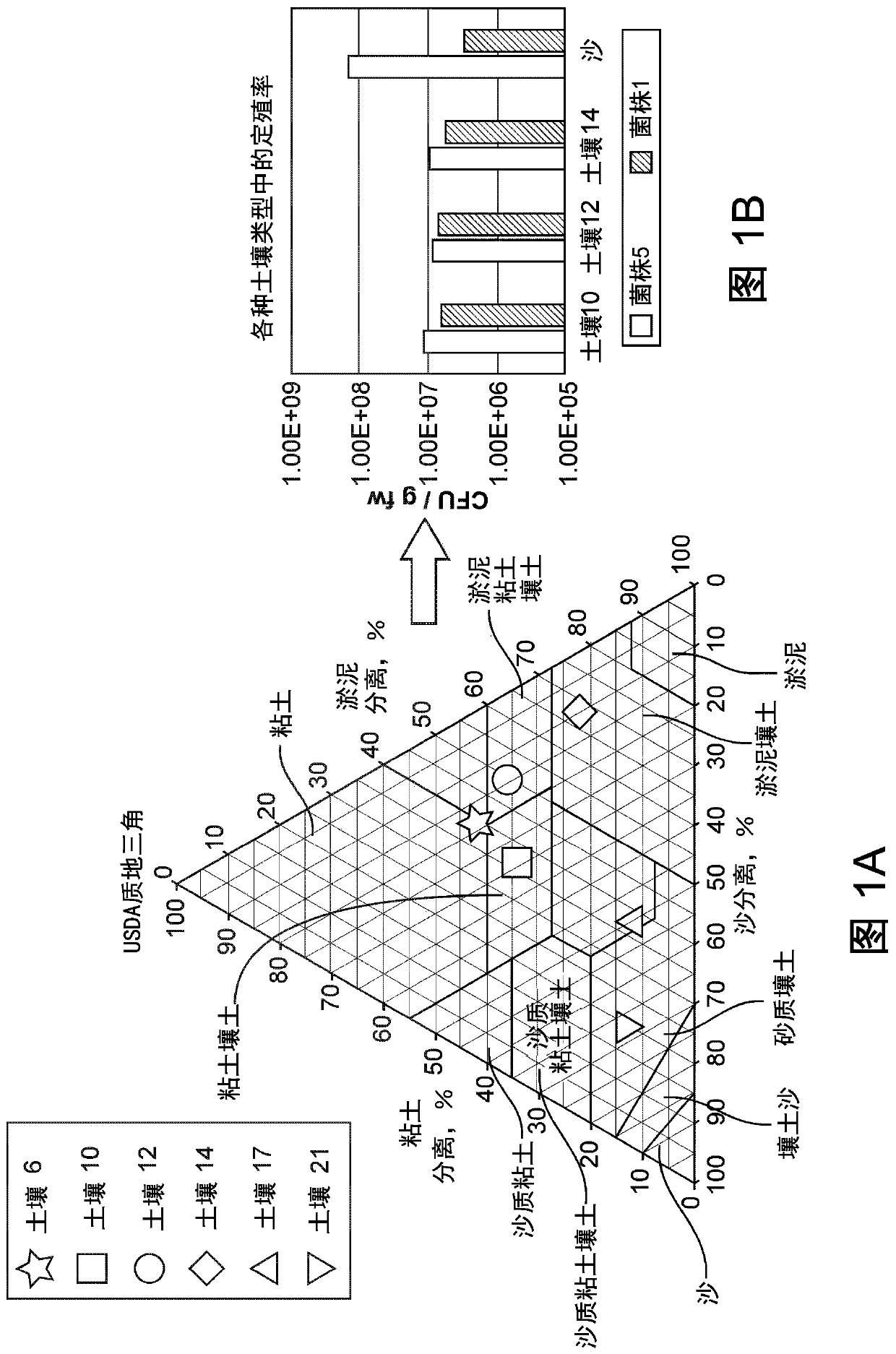
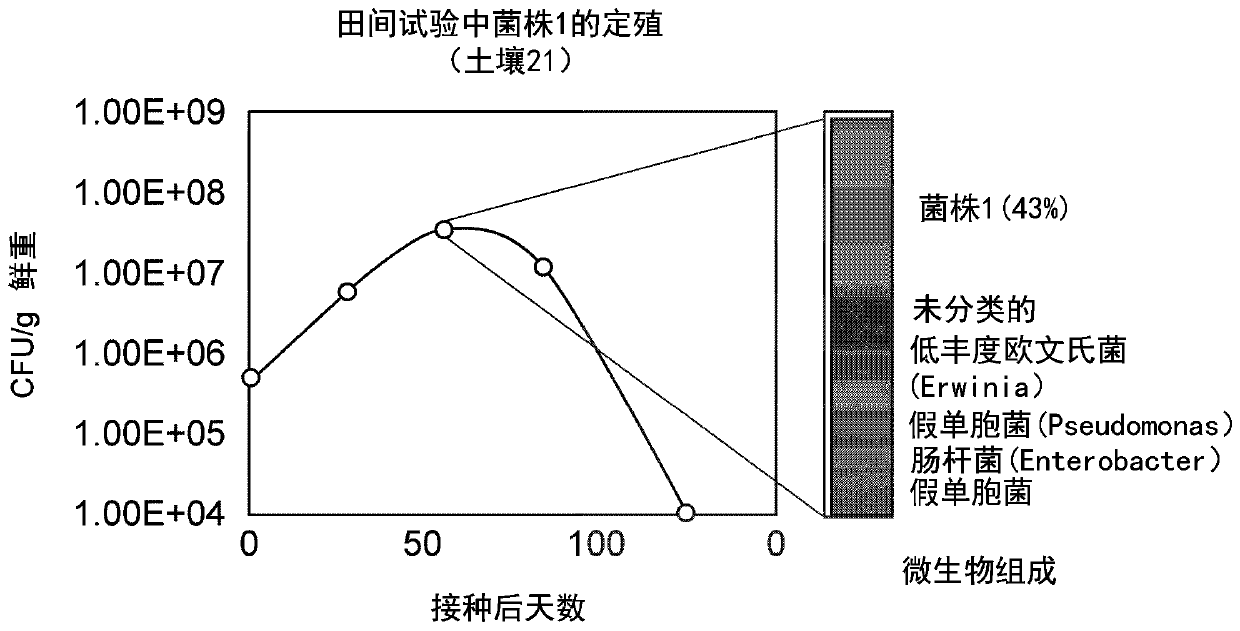
[0333] Colonization of plants by candidate microorganisms
[0334]Colonization of desired host plants by candidate microorganisms is quantified by short-term plant growth experiments. Maize plants were inoculated with strains expressing RFP from a plasmid or from a Tn5 integrated RFP expression cassette. Plants were grown in sterile sand and non-sterile peat media and inoculated by pipetting 1 mL of cell culture directly onto the coleoptiles of germinating plants three days after germination. The plasmid is maintained by watering the plants with a solution containing the appropriate antibiotic. After three weeks, plant roots were collected, rinsed three times in sterile water to remove visible soil, and divided into two samples. A root sample was analyzed by fluorescence microscopy to identify localization patterns of candidate microorganisms. Microscopic examination was performed on the finest intact plan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com