Application of a short DNA sequence in the identification of citrus taxa and citrus seedlings
A short-sequence, citrus technology, applied in the field of genetic engineering, can solve problems such as low overall cost, and achieve the effects of high specificity, high reliability and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
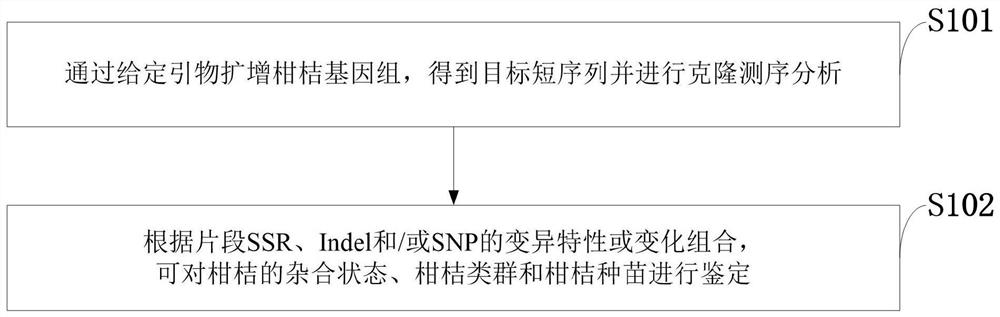
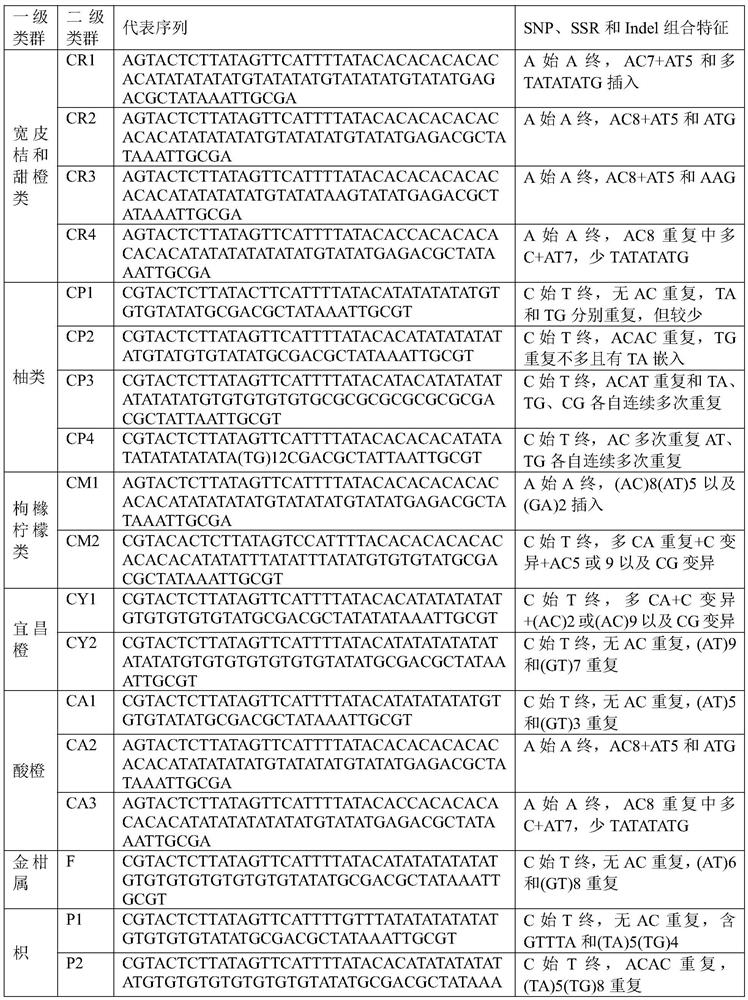
[0043] Example 1: Short sequence analysis of Wanjincheng
[0044] 1. The synthesis of forward and reverse primers used to amplify this short sequence requires acrylamide gel electrophoresis purification grade.
[0045] 2. Genomic DNA extraction of Wanjincheng. Genomic DNA of Wanjincheng was extracted by the improved CTAB method. The concentration of the extracted DNA was detected by a micro-volume ultraviolet spectrophotometer, the integrity of the extracted DNA was detected by 1% agarose gel electrophoresis, and it was stored at 4°C for future use.
[0046] 3. DNA polymerase chain reaction (PCR) amplification to obtain the target short sequence. The PCR system is as follows: 1×PCR Buffer, 1.5mmol L -1 Mg 2+ , 0.2mmol·L-1 of dNTPs, 0.33nmol·L -1 The upstream and downstream primers, 1U of Takara high-fidelity Taq DNA polymerase, about 100ng of DNA template, and a total volume of 50μL. The amplification program is: pre-denaturation at 94°C for 4 min; denaturation at 94°C f...
Embodiment 2
[0053] Example 2: Short sequence analysis of Oita Satsuma
[0054] 1. The synthesis of forward and reverse primers used to amplify this short sequence requires acrylamide gel electrophoresis purification grade.
[0055] 2. Genomic DNA extraction of Oita Satsuma. Genomic DNA of Oita Satsuma mandarin was extracted by the improved CTAB method. The concentration of the extracted DNA was detected by a micro-volume ultraviolet spectrophotometer, the integrity of the extracted DNA was detected by 1% agarose gel electrophoresis, and it was stored at 4°C for future use.
[0056] 3. DNA polymerase chain reaction (PCR) amplification to obtain the target short sequence. The PCR system is as follows: 1×PCR Buffer, 1.5mmol L -1 Mg 2+ , 0.2mmol·L-1 of dNTPs, 0.33nmol·L -1 The upstream and downstream primers, 1U of Takara high-fidelity Taq DNA polymerase, about 100ng of DNA template, and a total volume of 50μL. The amplification program is: pre-denaturation at 94°C for 4 min; denaturati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com