Chemically modified basic group-containing single-stranded DNA aptamer capable of specifically recognizing anthrax protective antigen PA83 and application thereof
A nucleic acid aptamer and DNA molecule technology, applied in the biological field, can solve the problems of being inferior to antibodies, the random base sequence should not be too long, limiting the amount of initial library information, etc., and achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1. Preparation of single-stranded DNA aptamers containing chemically modified bases that specifically recognize the anthrax protective antigen PA83
[0047] 1. Design and customize nucleic acid libraries of chemically modified nucleotides
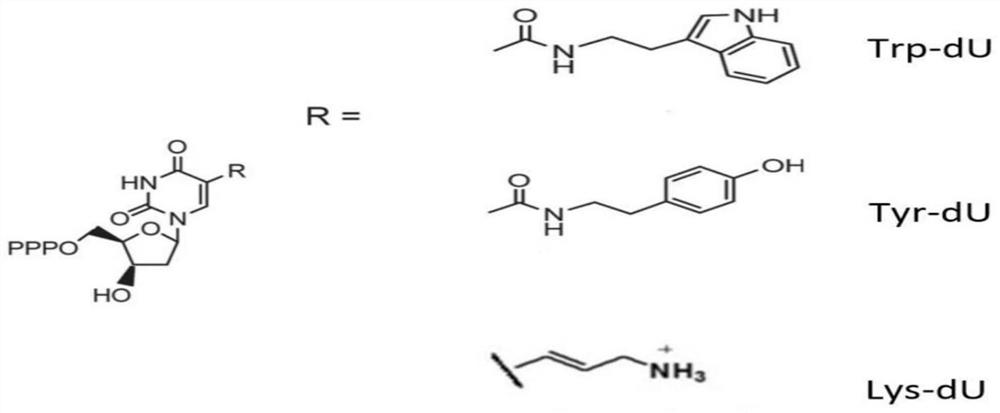
[0048] Because RNA aptamers are easily degraded by nucleases, the present invention selects and screens single-stranded DNA aptamers with stable structures in the detection environment. Select deoxyuracil (such as figure 1 shown) into the preparation of the nucleic acid library, respectively expressed as Trp-dU, Tyr-dU, Lys-dU. Indole groups, phenol groups and amine groups have the characteristics of nonpolar, polar uncharged and polar positively charged, respectively. The full-length nucleic acid library is 80bp, CAGGGGACGCACCAAGG-N40-CCATGACCCGCGTGCTGACATCG, the front and rear ends are fixed sequences for amplification, including only the natural nucleotide ATCG; the middle random sequences are designed to be 40, and synt...
Embodiment 2
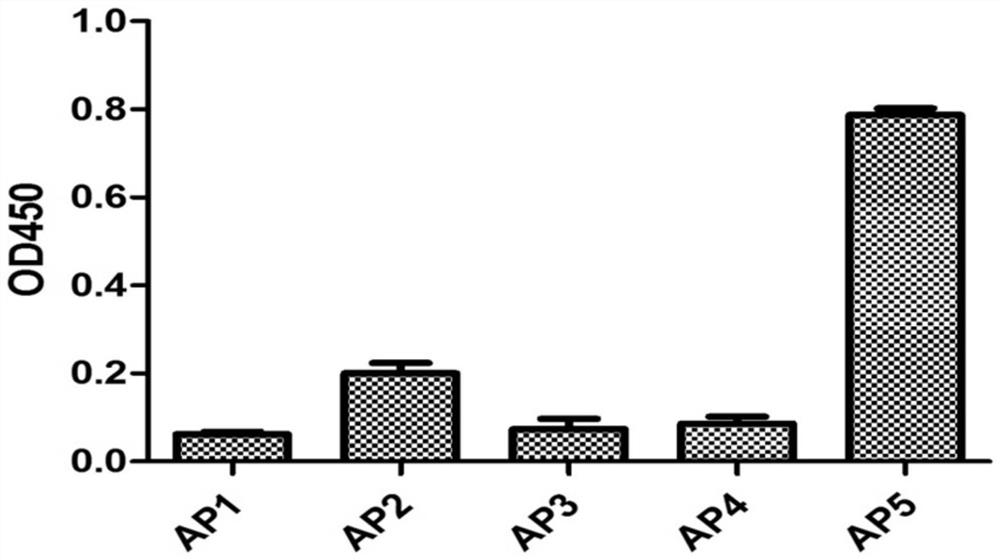
[0096] Example 2, the application of nucleic acid aptamer AP5 in the detection of PA83 protein
[0097] 1. Biotin-labeled nucleic acid aptamer AP5
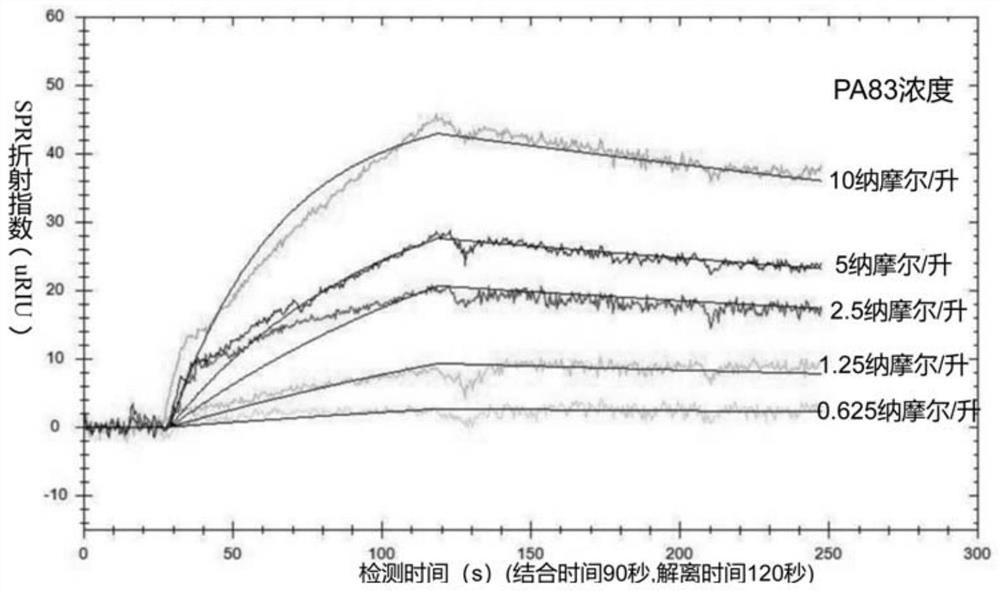
[0098] Synthesize biotin-labeled aptamer AP5, add a 10 dA sequence to the 5' end, 5'-Biotin-AAAAAAAAAA-AP5-3' (5'-Biotin-AAAAAAAAAAGCCCACGGCGGWYCGCCGGCCACAGTYAWCWGWGGTGGGC-3'), using RNAStructure software The secondary structure of the nucleic acid aptamer is predicted as Figure 4 .
[0099] 2. Construction of aptamer biosensor based on surface plasmon resonance technology by nucleic acid aptamer AP5
[0100] The method of constructing an aptamer biosensor based on surface plasmon resonance technology can refer to the following literature Wang S, Dong Y, Liang X. Development of a SPR aptasenxor containing oriented aptamer for direct capture and detection of tetracycline in multiple honey samples[J].Biosensors&Bioelectronics, 2018,109:1., the construction process is as follows:
PUM
| Property | Measurement | Unit |
|---|---|---|
| Full length | aaaaa | aaaaa |
| Equilibrium dissociation constant | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com