Methods and uses for promoting skeletal muscle development
A skeletal muscle and gene technology, applied in the field of promoting skeletal muscle development, can solve problems such as unclear regulatory mechanism, achieve wide applicability, promote skeletal muscle development, and promote development.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
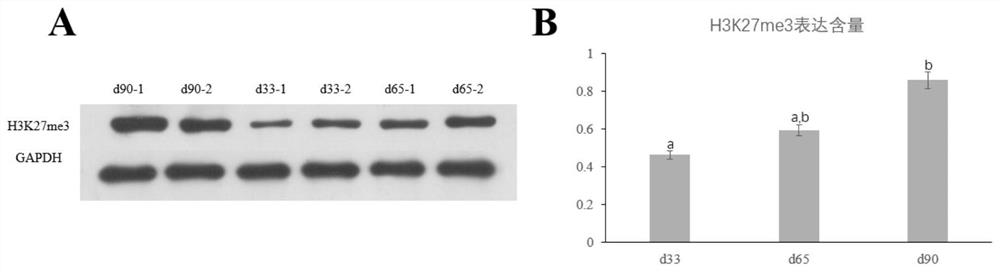
[0029] Example 1 Detection of H3K27me3 expression level in pig skeletal muscle samples in 3 different periods
[0030] A total of 6 tissue samples of the longissimus dorsi muscle samples of Duroc pig embryos at 33 days, 65 days and 90 days were taken for testing. The specific experimental process was as follows:
[0031] (1) Extraction of total protein
[0032] ① Prepare protein lysate.
[0033] ②Use clean scissors and tweezers to cut about 0.1g tissue sample, quickly put it into a pre-cooled homogenizer, and add protein lysate at a ratio of 1:10.
[0034] ③ Homogenize on ice until the tissue is digested, and transfer the sample to a 1.5mL centrifuge tube.
[0035] ④Centrifuge at 12,000×g at 4°C for 10 minutes, and transfer the supernatant to a new 1.5mL centrifuge tube.
[0036] (2) Protein quantification
[0037] According to the BCA protein quantification kit, the method is as follows:
[0038] ①Take 40μl protein standard (25mg / mL), add 1960μl PBS to dilute and prepare...
Embodiment 2
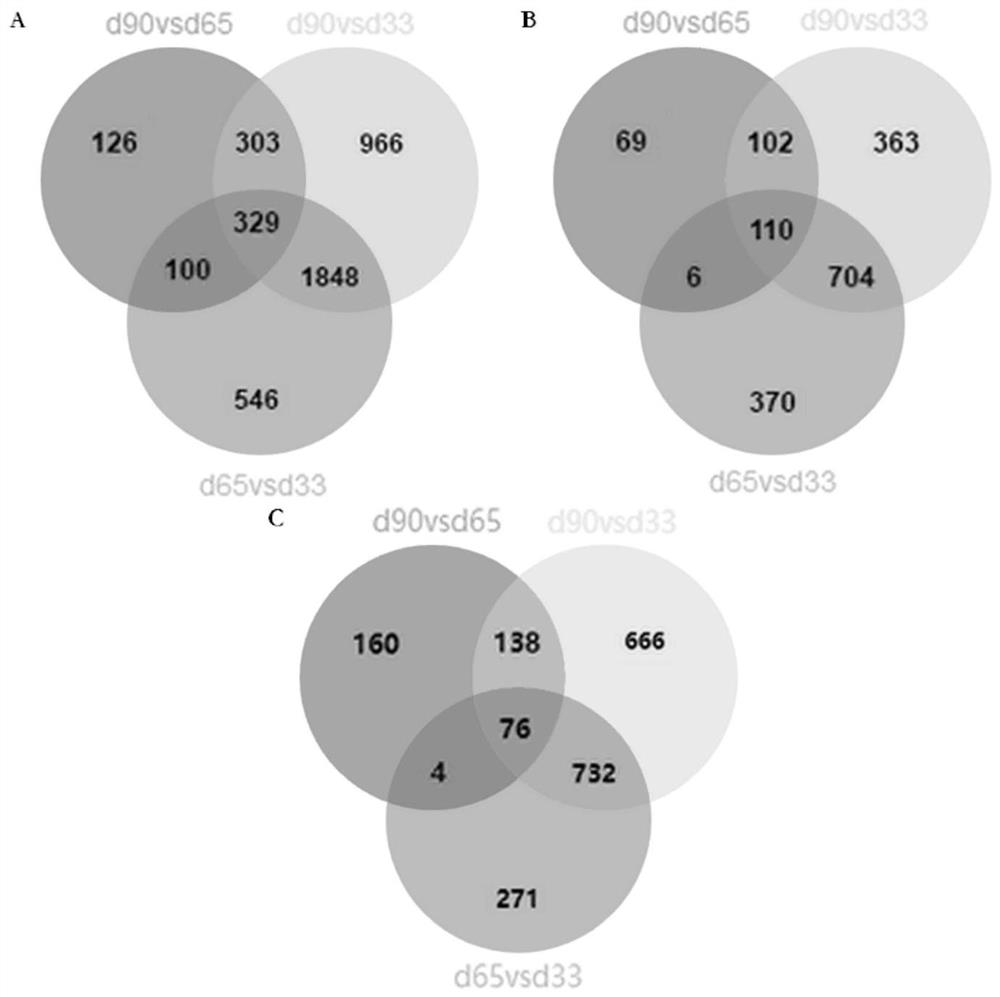
[0058] Example 2 Combined analysis of ChIP-seq and RNA-seq and screening of differentially expressed genes
[0059] 1. ChIP-seq experiment
[0060] (1) Preparation of working reagents
[0061] The working reagent preparation system used in the ChIP-seq experimental process is as follows:
[0062] ①Douncing Buffer (1mL): add 10μl Tris-HCl (pH=7.5), 4μl MgCl2, 2μl CaCl2, 10μl PIC, 974μl ddH2O respectively;
[0063] ②IP Buffer (1 mL): add 10 μl Tris-HCl (pH=8.0), 10 μl Triton X-100, 10 μl Deoxycholate (DOC), 10 μl SDS, 18 μl NaCl, 4 μl EDTA, 10 μl PIC, 928 μl ddH2O;
[0064] ③Hypotonic Lysis Buffer (1 mL): add 0.4 μl EDTA (pH=8.0), 0.1 μl Benzamidine, 1 μl PMSF, 3 μl DTT, 10 μl PIC, 985.5 μl ddH2O respectively;
[0065] ④ChiP Wash Buffer (1mL): add 20μl Tris-HCl (pH=8.0), 10μl SDS, 10μl Triton X-100, 4μl EDTA, 30μl NaCl, 10μl PIC, 916μl ddH2O respectively;
[0066] ⑤ChiP Final Wash Buffer (1mL): add 20μl Tris-HCl (pH=8.0), 10μl SDS, 10μl Triton X-100, 4μl EDTA, 100μl NaCl, 10...
Embodiment 3
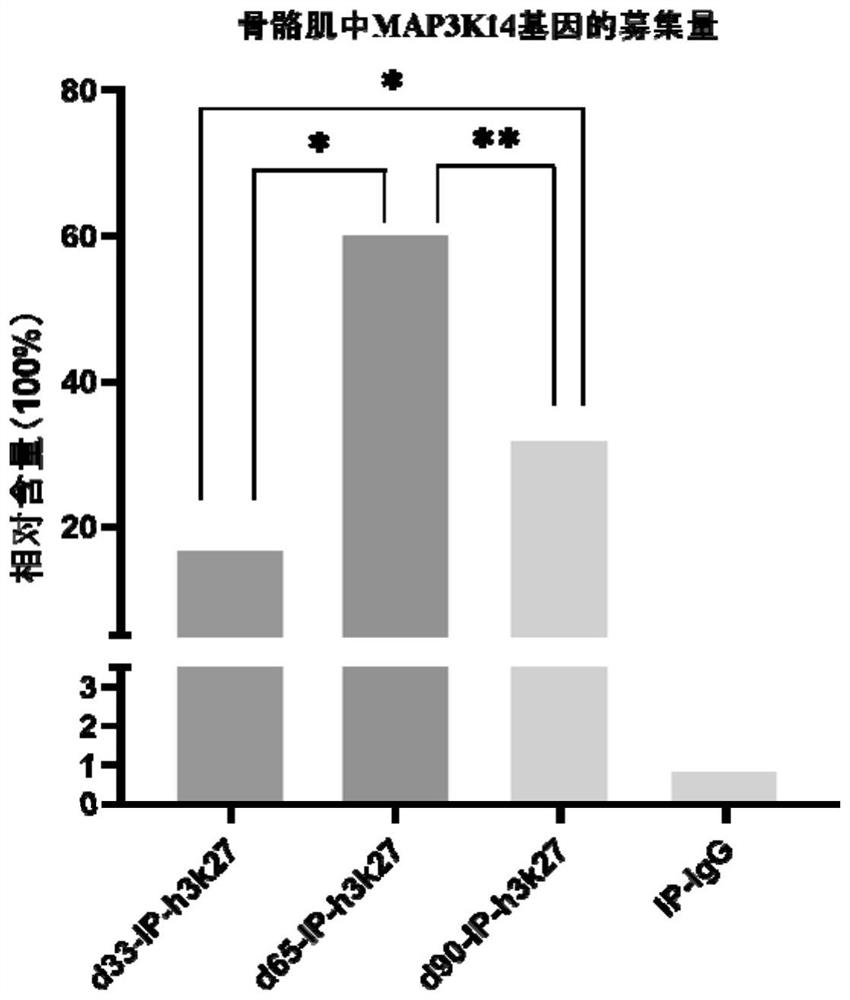
[0157] Example 3 ChIP-qPCR verification of differentially expressed gene MAP3K14
[0158] The quantitative PCR primers for porcine genes were designed using the primer design software Oligo 7.56 as shown in Table 2. Dilute the IP samples incubated with antibodies obtained during chromatin immunoprecipitation to the same concentration, and dilute the Input of 9 samples to 10ng / mL, 5ng / mL, 2.5ng / mL, 1.25ng / mL, 0.625 ng / mL, different concentrations of Input were used to draw a standard curve, and all samples were prepared according to Table 3. Amplification system. Absolute quantitative PCR was performed using illumina's EcoTM Real-time PCR instrument. Each IP sample and different concentrations of Input were used for 3 biological repetitions. The reaction conditions were: 95°C, 5min; 95°C, 10s, 60°C, 15s, 72 ℃, 20s, 40-50 cycles; 95℃, 15s; 60℃, 15s; 95℃, 15s.
[0159] Table 2 ChIP-qPCR primers
[0160]
[0161]Table 3 ChIP-qPCR reaction system
[0162]
[0163] The res...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com