Recombinant canine measles virus expressing luciferase
A technology of measles virus and enzyme cleavage site, which can be applied to viruses, viral peptides, viruses/phages, etc., and can solve problems such as high-efficiency expression of NLuc
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Embodiment 1, construction of rCMV-NLuc cDNAclone
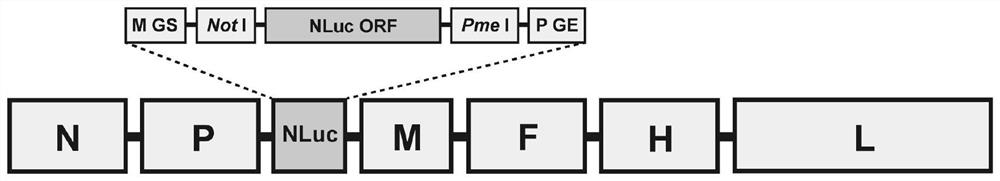
[0025] The recombinant CMV nucleic acid fragment rCMV-NLuc cDNA clone used to rescue and express NLuc constructed in this example was distributed in "5'-N-P-NLuc-M-F-H-L-3'" ( figure 1 ); wherein, N, P, M, F, H, L are structural genes of the virus; the selected N, P, M, F, H, L genes of this embodiment are CMV strain 5804P (GenBank: AY386316.1) related genes; NLuc is The ORF of luciferase, the upstream of the ORF includes MGS and Not I restriction sites, and the downstream includes Pme I restriction sites and PGE.
[0026] In order to obtain an ideal rescue efficiency, the present invention inserts the exogenous NLuc gene between the P and M genes; at the same time, in order to increase the expression of NLuc, a Kozak sequence is inserted upstream of the NLuc ORF.
[0027] The nucleotide sequence of one rCMV-NLuc cDNA clone is SEQ ID NO:1.
[0028] The full-length sequence of the rCMV-NLuc cDNA clone whose nucleoti...
Embodiment 2
[0029] Embodiment 2, the rescue of rCMV-NLuc
[0030] Resuscitate BSR-T7 / 5 cells, passage twice in DMEM medium, DMEM medium contains penicillin (100U / mL), streptomycin (100μg / mL), amphotericin B (0.25μg / mL), G418 (500μg / mL) And 10% FBS (fetal bovine serum). The day before transfection, inoculate BSR-T7 / 5 cells in a 6-well plate, and when the cells reach 70% confluence the next day, clone rCMV-NLuc cDNA clone plasmids and eukaryotic expression plasmids expressing CMV N, P, and L proteins respectively BSR-T7 / 5 cells were co-transfected, and the transfection amounts of the four methods were 4 μg, 2 μg, 1 μg, and 1 μg per well, respectively. Prepare Solution I by adding 150 μL Opti-MEM and 16 μL Lipofectamine 2000 to each well. Add 150 μL Opti-MEM and 8 μg plasmid to each well to prepare solution II. Gently mix solutions I and II, and place at room temperature for 15 minutes to form liposome-DNA complexes. Aspirate the culture medium of the 6-well plate, add 1mL Opti-MEM to ea...
Embodiment 3
[0031] Embodiment 3, blind transmission and identification of rCMV-NLuc
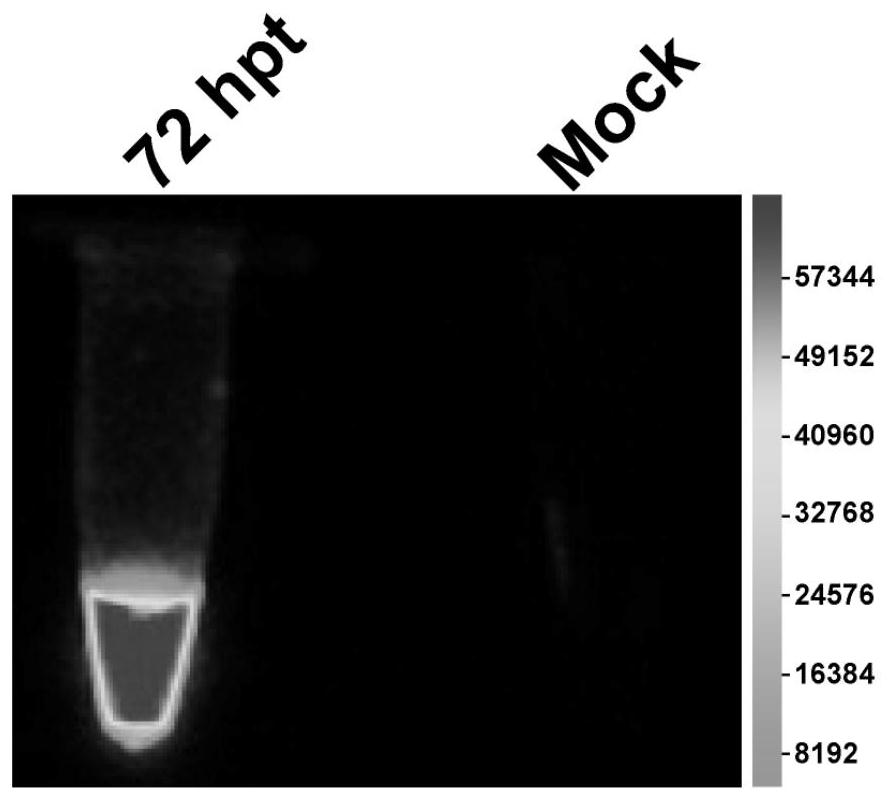
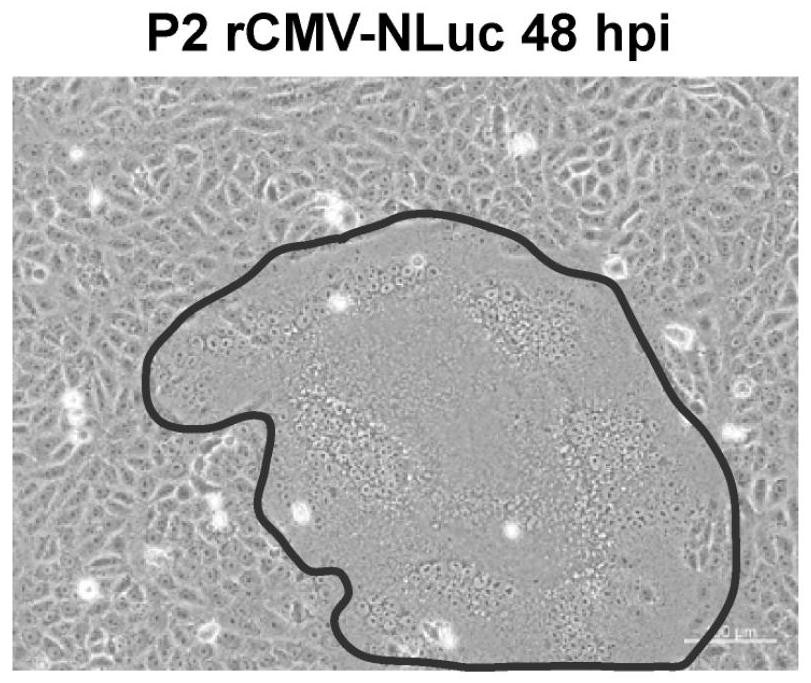
[0032] The rescued rCMV-NLuc was blindly passed in VDS cells, and syncytial cell lesions could be observed in the second generation (P2) ( image 3 ). Continue to blindly pass to the fifth generation (P5), the virus solution is frozen and thawed once, and the supernatant is collected to extract total RNA, which is used for RT-PCR detection of the virus. The upstream primer sequence: 5'-gatcaaaagtatcacacatgcttaa-3', the downstream primer sequence: 5'-gatcgaagtcgtacacctcagtcat-3', amplifying a 799bp fragment. The RT-PCR reaction used High Fidelity OneStep RT-PCR Kit (Takara), and the reaction conditions were set as follows: 45°C for 10 minutes, 94°C for 2 minutes, and 30 cycles: 98°C (10s), 55°C (15s), 68°C ( 10s). In order to eliminate residual plasmid contamination, another PCR control was set up. The PCR reaction used 2×PrimeSTAR Max Premix (Takara), and the reaction conditions were set to 30 cycles...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com